Genetic control of the innate immune response
- PMID: 12826024
- PMCID: PMC194878
- DOI: 10.1186/1471-2172-4-5
Genetic control of the innate immune response
Abstract
Background: Susceptibility to infectious diseases is directed, in part, by the interaction between the invading pathogen and host macrophages. This study examines the influence of genetic background on host-pathogen interactions, by assessing the transcriptional responses of macrophages from five inbred mouse strains to lipopolysaccharide (LPS), a major determinant of responses to gram-negative microorganisms.
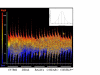
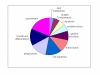
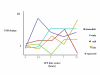
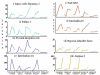
Results: The mouse strains examined varied greatly in the number, amplitude and rate of induction of genes expressed in response to LPS. The response was attenuated in the C3H/HeJlpsd strain, which has a mutation in the LPS receptor Toll-like receptor 4 (TLR4). Variation between mouse strains allowed clustering into early (C57Bl/6J and DBA/2J) and delayed (BALB/c and C3H/ARC) transcriptional phenotypes. There was no clear correlation between gene induction patterns and variation at the Bcg locus (Slc11A1) or propensity to bias Th1 versus Th2 T cell activation responses.
Conclusion: Macrophages from each strain responded to LPS with unique gene expression profiles. The variation apparent between genetic backgrounds provides insights into the breadth of possible inflammatory responses, and paradoxically, this divergence was used to identify a common transcriptional program that responds to TLR4 signalling, irrespective of genetic background. Our data indicates that many additional genetic loci control the nature and the extent of transcriptional responses promoted by a single pathogen-associated molecular pattern (PAMP), such as LPS.
Figures


Similar articles
-
Positional cloning of Lps, and the general role of toll-like receptors in the innate immune response.Eur Cytokine Netw. 2000 Jun;11(2):143-52. Eur Cytokine Netw. 2000. PMID: 10903793 Review.
-
Upregulation of toll-like receptor 2 gene expression in macrophage response to peptidoglycan and high concentration of lipopolysaccharide is involved in NF-kappa b activation.Infect Immun. 2001 May;69(5):2788-96. doi: 10.1128/IAI.69.5.2788-2796.2001. Infect Immun. 2001. PMID: 11292690 Free PMC article.
-
Toll-like receptor-mediated activation of B cells and macrophages by polysaccharide isolated from cell culture of Acanthopanax senticosus.Int Immunopharmacol. 2003 Sep;3(9):1301-12. doi: 10.1016/S1567-5769(03)00118-8. Int Immunopharmacol. 2003. PMID: 12890428
-
The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors.Nature. 2002 Nov 21;420(6913):329-33. doi: 10.1038/nature01180. Nature. 2002. PMID: 12447442
-
Receptors for innate pathogen defence in insects are normal activation receptors for specific immune responses in mammals.Scand J Immunol. 1999 Oct;50(4):341-7. doi: 10.1046/j.1365-3083.1999.00605.x. Scand J Immunol. 1999. PMID: 10520172 Review.
Cited by
-
Pulmonary responses to Stachybotrys chartarum and its toxins: mouse strain affects clearance and macrophage cytotoxicity.Toxicol Sci. 2010 Jul;116(1):113-21. doi: 10.1093/toxsci/kfq104. Epub 2010 Apr 12. Toxicol Sci. 2010. PMID: 20385656 Free PMC article.
-
A comparative encyclopedia of DNA elements in the mouse genome.Nature. 2014 Nov 20;515(7527):355-64. doi: 10.1038/nature13992. Nature. 2014. PMID: 25409824 Free PMC article.
-
Functional plasticity of macrophages: reversible adaptation to changing microenvironments.J Leukoc Biol. 2004 Sep;76(3):509-13. doi: 10.1189/jlb.0504272. Epub 2004 Jun 24. J Leukoc Biol. 2004. PMID: 15218057 Free PMC article. Review.
-
The Interplay between Salmonella and Intestinal Innate Immune Cells in Chickens.Pathogens. 2021 Nov 19;10(11):1512. doi: 10.3390/pathogens10111512. Pathogens. 2021. PMID: 34832668 Free PMC article. Review.
-
Host genetic background impacts modulation of the TLR4 pathway by RON in tissue-associated macrophages.Immunol Cell Biol. 2013 Aug;91(7):451-60. doi: 10.1038/icb.2013.27. Epub 2013 Jul 2. Immunol Cell Biol. 2013. PMID: 23817579 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

