Many paths to methyltransfer: a chronicle of convergence
- PMID: 12826405
- PMCID: PMC2758044
- DOI: 10.1016/S0968-0004(03)00090-2
Many paths to methyltransfer: a chronicle of convergence
Abstract
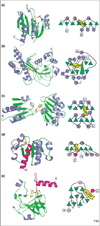
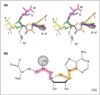
S-adenosyl-L-methionine (AdoMet) dependent methyltransferases (MTases) are involved in biosynthesis, signal transduction, protein repair, chromatin regulation and gene silencing. Five different structural folds (I-V) have been described that bind AdoMet and catalyze methyltransfer to diverse substrates, although the great majority of known MTases have the Class I fold. Even within a particular MTase class the amino-acid sequence similarity can be as low as 10%. Thus, the structural and catalytic requirements for methyltransfer from AdoMet appear to be remarkably flexible.
Figures


References
-
- Cantoni GL. Biological methylation: selected aspects. Annu. Rev. Biochem. 1975;44:435–451. - PubMed
-
- Ohsawa N, et al. Purification and characterization of a monohalomethane-producing enzyme S-adenosyl-l-methionine: halide ion methyltransferase from a marine microalga, Pavlova pinguis. Biosci. Biotechnol. Biochem. 2001;65:2397–2404. - PubMed
-
- Attieh JM, et al. Purification and characterization of a novel methyltransferase responsible for biosynthesis of halomethanes and methanethiol in Brassica oleracea. J. Biol. Chem. 1995;270:9250–9257. - PubMed
-
- Cheng X, et al. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. - PubMed
-
- Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

