The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants
- PMID: 12826610
- PMCID: PMC166205
- DOI: 10.1073/pnas.1431624100
The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants
Abstract
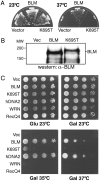
Bloom syndrome is a disorder of profound and early cancer predisposition in which cells become hypermutable, exhibit high frequency of sister chromatid exchanges, and show increased micronuclei. BLM, the gene mutated in Bloom syndrome, has been cloned previously, and the BLM protein is a member of the RecQ family of DNA helicases. Many lines of evidence suggest that BLM is involved either directly in DNA replication or in surveillance during DNA replication, but its specific roles remain unknown. Here we show that hBLM can suppress both the temperature-sensitive growth defect and the DNA damage sensitivity of the yeast DNA replication mutant dna2-1. The dna2-1 mutant is defective in a helicase-nuclease that is required either to coordinate with the crucial Saccharomyces cerevisiae (sc) FEN1 nuclease in Okazaki fragment maturation or to compensate for scFEN1 when its activity is impaired. We show that human BLM interacts with both scDna2 and scFEN1 by using coimmunoprecipitation from yeast extracts, suggesting that human BLM participates in the same steps of DNA replication or repair as scFEN1 and scDna2.
Figures




Similar articles
-
RecQ helicases: caretakers of the genome.Nat Rev Cancer. 2003 Mar;3(3):169-78. doi: 10.1038/nrc1012. Nat Rev Cancer. 2003. PMID: 12612652 Review.
-
Werner and Bloom helicases are involved in DNA repair in a complementary fashion.Oncogene. 2002 Jan 31;21(6):954-63. doi: 10.1038/sj.onc.1205143. Oncogene. 2002. PMID: 11840341
-
In vivo function of the conserved non-catalytic domain of Werner syndrome helicase in DNA replication.Hum Mol Genet. 2004 Oct 1;13(19):2247-61. doi: 10.1093/hmg/ddh234. Epub 2004 Jul 28. Hum Mol Genet. 2004. PMID: 15282207
-
The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways.Mutat Res. 2000 Apr 28;459(3):173-86. doi: 10.1016/s0921-8777(99)00072-5. Mutat Res. 2000. PMID: 10812329
-
[Function of RecQ family helicases and Bloom's syndrome].Tanpakushitsu Kakusan Koso. 2001 Jun;46(8 Suppl):1082-8. Tanpakushitsu Kakusan Koso. 2001. PMID: 11436296 Review. Japanese. No abstract available.
Cited by
-
An enhanced CRISPR repressor for targeted mammalian gene regulation.Nat Methods. 2018 Aug;15(8):611-616. doi: 10.1038/s41592-018-0048-5. Epub 2018 Jul 16. Nat Methods. 2018. PMID: 30013045 Free PMC article.
-
A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination.Mol Cell. 2015 Aug 6;59(3):478-90. doi: 10.1016/j.molcel.2015.07.009. Mol Cell. 2015. PMID: 26253028 Free PMC article.
-
The wonders of flap endonucleases: structure, function, mechanism and regulation.Subcell Biochem. 2012;62:301-26. doi: 10.1007/978-94-007-4572-8_16. Subcell Biochem. 2012. PMID: 22918592 Free PMC article. Review.
-
Okazaki fragment processing-independent role for human Dna2 enzyme during DNA replication.J Biol Chem. 2012 Jun 22;287(26):21980-91. doi: 10.1074/jbc.M112.359018. Epub 2012 May 7. J Biol Chem. 2012. PMID: 22570476 Free PMC article.
-
Multiple roles of DNA2 nuclease/helicase in DNA metabolism, genome stability and human diseases.Nucleic Acids Res. 2020 Jan 10;48(1):16-35. doi: 10.1093/nar/gkz1101. Nucleic Acids Res. 2020. PMID: 31754720 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

