Modulation of early olfactory processing by an octopaminergic reinforcement pathway in the honeybee
- PMID: 12832563
- PMCID: PMC6741157
- DOI: 10.1523/JNEUROSCI.23-12-05370.2003
Modulation of early olfactory processing by an octopaminergic reinforcement pathway in the honeybee
Abstract
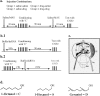
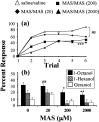
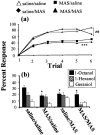
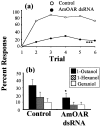
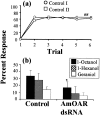
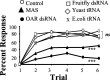
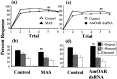
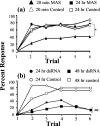
Processing of olfactory information in the antennal lobes of insects and olfactory bulbs of vertebrates is modulated by centrifugal inputs that represent reinforcing events. Octopamine release by one such pathway in the honeybee antennal lobe modulates olfactory processing in relation to nectar (sucrose) reinforcement. To test more specifically what role octopamine plays in the antennal lobe, we used two treatments to disrupt an octopamine receptor from Apis mellifera brain (AmOAR) function: (1) an OAR antagonist, mianserin, was used to block receptor function, and (2) AmOAR double-stranded RNA was used to silence receptor expression. Both treatments inhibited olfactory acquisition and recall, but they did not disrupt odor discrimination. These results suggest that octopamine mediates consolidation of a component of olfactory memory at this early processing stage in the antennal lobe. Furthermore, after consolidation, octopamine release becomes essential for recall, which suggests that the modulatory circuits become incorporated as essential components of neural representations that activate odor memory.
Figures


References
-
- Billuart P, Winter CG, Maresh A, Zhao X, Luo L ( 2001) Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell 107: 195–207. - PubMed
-
- Bitterman ME, Menzel R, Fietz A, Schafer S ( 1983) Classical conditioning of proboscis extension in honeybees (Apis mellifera). J Comp Physiol 97: 107–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
