Annotation and evolutionary relationships of a small regulatory RNA gene micF and its target ompF in Yersinia species
- PMID: 12834539
- PMCID: PMC166144
- DOI: 10.1186/1471-2180-3-13
Annotation and evolutionary relationships of a small regulatory RNA gene micF and its target ompF in Yersinia species
Abstract
Background: micF RNA, a small regulatory RNA found in bacteria, post-transcriptionally regulates expression of outer membrane protein F (OmpF) by interaction with the ompF mRNA 5'UTR. Phylogenetic data can be useful for RNA/RNA duplex structure analyses and aid in elucidation of mechanism of regulation. However micF and associated genes, ompF and ompC are difficult to annotate because of either similarities or divergences in nucleotide sequence. We report by using sequences that represent "gene signatures" as probes, e.g., mRNA 5'UTR sequences, closely related genes can be accurately located in genomic sequences.
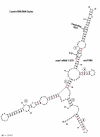
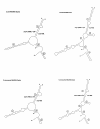
Results: Alignment and search methods using NCBI BLAST programs have been used to identify micF, ompF and ompC in Yersinia pestis and Yersinia enterocolitica. By alignment with DNA sequences from other bacterial species, 5' start sites of genes and upstream transcriptional regulatory sites in promoter regions were predicted. Annotated genes from Yersinia species provide phylogenetic information on the micF regulatory system. High sequence conservation in binding sites of transcriptional regulatory factors are found in the promoter region upstream of micF and conservation in blocks of sequences as well as marked sequence variation is seen in segments of the micF RNA gene. Unexpected large differences in rates of evolution were found between the interacting RNA transcripts, micF RNA and the 5' UTR of the ompF mRNA. micF RNA/ompF mRNA 5' UTR duplex structures were modeled by the mfold program. Functional domains such as RNA/RNA interacting sites appear to display a minimum of evolutionary drift in sequence with the exception of a significant change in Y. enterocolitica micF RNA.
Conclusions: Newly annotated Yersinia micF and ompF genes and the resultant RNA/RNA duplex structures add strong phylogenetic support for a generalized duplex model. The alignment and search approach using 5' UTR signatures may be a model to help define other genes and their start sites when annotated genes are available in well-defined reference organisms.
Figures






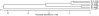



Similar articles
-
Antisense micF RNA and 5'-UTR of the target ompF RNA: phylogenetic conservation of primary and secondary structures.Nucleic Acids Symp Ser. 1997;(36):33-5. Nucleic Acids Symp Ser. 1997. PMID: 9478198
-
Secondary structures of Escherichia coli antisense micF RNA, the 5'-end of the target ompF mRNA, and the RNA/RNA duplex.Biochemistry. 1995 Mar 21;34(11):3621-31. doi: 10.1021/bi00011a017. Biochemistry. 1995. PMID: 7534474
-
Intrinsic plasmids influence MicF-mediated translational repression of ompF in Yersinia pestis.Front Microbiol. 2015 Aug 21;6:862. doi: 10.3389/fmicb.2015.00862. eCollection 2015. Front Microbiol. 2015. PMID: 26347736 Free PMC article.
-
MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors.J Mol Biol. 2001 Oct 12;313(1):1-12. doi: 10.1006/jmbi.2001.5029. J Mol Biol. 2001. PMID: 11601842 Review.
-
Phylogenetic analysis of general bacterial porins: a phylogenomic case study.J Mol Microbiol Biotechnol. 2006;11(6):291-301. doi: 10.1159/000095631. J Mol Microbiol Biotechnol. 2006. PMID: 17114893 Review.
Cited by
-
Molecular Evolution of the Yersinia Major Outer Membrane Protein C (OmpC).Evol Bioinform Online. 2016 Aug 21;12:185-91. doi: 10.4137/EBO.S40346. eCollection 2016. Evol Bioinform Online. 2016. PMID: 27578962 Free PMC article.
-
IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions.Bioinformatics. 2008 Dec 15;24(24):2849-56. doi: 10.1093/bioinformatics/btn544. Epub 2008 Oct 21. Bioinformatics. 2008. PMID: 18940824 Free PMC article.
-
Evolutionary patterns of non-coding RNAs.Theory Biosci. 2005 Apr;123(4):301-69. doi: 10.1016/j.thbio.2005.01.002. Theory Biosci. 2005. PMID: 18202870
-
Post-transcriptional regulation of gene expression in Yersinia species.Front Cell Infect Microbiol. 2012 Nov 9;2:129. doi: 10.3389/fcimb.2012.00129. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 23162797 Free PMC article. Review.
-
Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Yersinia pestis.BMC Microbiol. 2011 Feb 23;11:39. doi: 10.1186/1471-2180-11-39. BMC Microbiol. 2011. PMID: 21345178 Free PMC article.
References
-
- Schmidt M, Zheng P, Delihas N. Secondary structures of Escherichia coli antisense micF RNA, the 5'-end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry. 1995;34:3621–3631. - PubMed
-
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder NJ, Oinn TM, Pagni M, Servant F, Sigrist CJ, Zdobnov EM, InterPro Consortium. InterPro – an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. doi: 10.1093/bioinformatics/16.12.1145. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

