Complete genome sequence of the marine planctomycete Pirellula sp. strain 1
- PMID: 12835416
- PMCID: PMC166223
- DOI: 10.1073/pnas.1431443100
Complete genome sequence of the marine planctomycete Pirellula sp. strain 1
Abstract
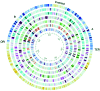
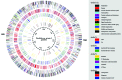
Pirellula sp. strain 1 ("Rhodopirellula baltica") is a marine representative of the globally distributed and environmentally important bacterial order Planctomycetales. Here we report the complete genome sequence of a member of this independent phylum. With 7.145 megabases, Pirellula sp. strain 1 has the largest circular bacterial genome sequenced so far. The presence of all genes required for heterolactic acid fermentation, key genes for the interconversion of C1 compounds, and 110 sulfatases were unexpected for this aerobic heterotrophic isolate. Although Pirellula sp. strain 1 has a proteinaceous cell wall, remnants of genes for peptidoglycan synthesis were found. Genes for lipid A biosynthesis and homologues to the flagellar L- and P-ring protein indicate a former Gram-negative type of cell wall. Phylogenetic analysis of all relevant markers clearly affiliates the Planctomycetales to the domain Bacteria as a distinct phylum, but a deepest branching is not supported by our analyses.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

