Crystal structure of the YchF protein reveals binding sites for GTP and nucleic acid
- PMID: 12837776
- PMCID: PMC164861
- DOI: 10.1128/JB.185.14.4031-4037.2003
Crystal structure of the YchF protein reveals binding sites for GTP and nucleic acid
Abstract
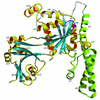
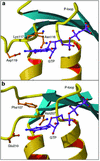
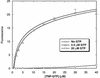
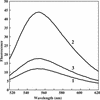
The bacterial protein encoded by the gene ychF is 1 of 11 universally conserved GTPases and the only one whose function is unknown. The crystal structure determination of YchF was sought to help with the functional assignment of the protein. The YchF protein from Haemophilus influenzae was cloned and expressed, and the crystal structure was determined at 2.4 A resolution. The polypeptide chain is folded into three domains. The N-terminal domain has a mononucleotide binding fold typical for the P-loop NTPases. An 80-residue domain next to it has a pronounced alpha-helical coiled coil. The C-terminal domain features a six-stranded half-barrel that curves around an alpha-helix. The crablike three-domain structure of YchF suggests the binding site for a double-stranded nucleic acid in the cleft between the domains. The structure of the putative GTP-binding site is consistent with the postulated guanine specificity of the protein. Fluorescence measurements have demonstrated the ability of YchF to bind a double-stranded nucleic acid and GTP. Taken together with other experimental data and genomic analysis, these results suggest that YchF may be part of a nucleoprotein complex and may function as a GTP-dependent translation factor.
Figures


References
-
- Biou, V., A. Yaremchuk, M. Tukalo, and S. Cusack. 1994. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263:1404-1410. - PubMed
-
- Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348:125-132. - PubMed
-
- Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

