A novel adenylate binding site confers phosphopantetheine adenylyltransferase interactions with coenzyme A
- PMID: 12837781
- PMCID: PMC164871
- DOI: 10.1128/JB.185.14.4074-4080.2003
A novel adenylate binding site confers phosphopantetheine adenylyltransferase interactions with coenzyme A
Abstract
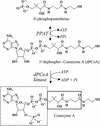
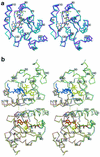
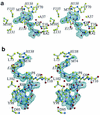
Phosphopantetheine adenylyltransferase (PPAT) regulates the key penultimate step in the essential coenzyme A (CoA) biosynthetic pathway. PPAT catalyzes the reversible transfer of an adenylyl group from Mg(2+):ATP to 4'-phosphopantetheine to form 3'-dephospho-CoA (dPCoA) and pyrophosphate. The high-resolution crystal structure of PPAT complexed with CoA has been determined. Remarkably, CoA and the product dPCoA bind to the active site in distinct ways. Although the phosphate moiety within the phosphopantetheine arm overlaps, the pantetheine arm binds to the same pocket in two distinct conformations, and the adenylyl moieties of these two ligands have distinct binding sites. Moreover, the PPAT:CoA crystal structure confirms the asymmetry of binding to the two trimers within the hexameric enzyme. Specifically, the pantetheine arm of CoA bound to one protomer within the asymmetric unit displays the dPCoA-like conformation with the adenylyl moiety disordered, whereas CoA binds the twofold-related protomer in an ordered and unique fashion.
Figures
References
-
- Bork, P., L. Holm, E. V. Koonin, and C. Sander. 1995. The cytidylyltransferase superfamily: identification of the nucleotide-binding site and fold prediction. Proteins 22:259-266. - PubMed
-
- Brünger, A. T., J. Kuriyan, and M. Karplus. 1988. Crystallographic R-factor refinement by molecular dynamics. Science 235:458-460. - PubMed
-
- Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kunszeweski, N. Nilges, M. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. - PubMed
-
- Brünger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

