Chimeric recombinases with designed DNA sequence recognition
- PMID: 12837939
- PMCID: PMC166373
- DOI: 10.1073/pnas.1533177100
Chimeric recombinases with designed DNA sequence recognition
Abstract
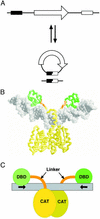
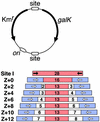
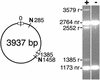
Site-specific recombination typically occurs only between DNA sequences that have co-evolved with a natural recombinase enzyme to optimize sequence recognition, catalytic efficiency, and regulation. Here, we show that the sequence recognition and the catalysis functions of a recombinase can be specified by unrelated protein domains. We describe chimeric recombinases with a catalytic domain from an activated multiple mutant of the bacterial enzyme Tn3 resolvase, fused to a DNA recognition domain from the mouse transcription factor Zif268. These proteins catalyze efficient recombination specifically at synthetic target sites recognized by two Zif268 domains. Our results demonstrate the functional autonomy of the resolvase catalytic domain and open the way to creating "custom-built" recombinases that act at chosen natural target sequences.
Figures

References
-
- Nash, H. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), 2nd Ed., Vol. 1, pp. 2363–2376.
-
- Gorman, C. & Bullock, C. (2000) Curr. Opin. Biotechnol. 11, 455–460. - PubMed
-
- Nagy, A. (2000) Genesis 26, 99–109. - PubMed
-
- Buchholz, F. & Stewart, A. F. (2001) Nat. Biotechnol. 19, 1047–1052. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

