Detection and diversity assessment of Xylella fastidiosa in field-collected plant and insect samples by using 16S rRNA and gyrB sequences
- PMID: 12839807
- PMCID: PMC165181
- DOI: 10.1128/AEM.69.7.4249-4255.2003
Detection and diversity assessment of Xylella fastidiosa in field-collected plant and insect samples by using 16S rRNA and gyrB sequences
Abstract
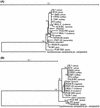
The causal agent of diseases in many economically important plants is attributed to the xylem-limited bacterium Xylella fastidiosa. The detection of this plant pathogen has been hampered due to its difficult isolation and slow growth on plates. Nearly complete nucleotide sequences of the 16S rRNA gene and partial sequences of the gyrB gene were determined for 18 strains of X. fastidiosa isolated from different plant hosts. A phylogenetic analysis, based on gyrB, grouped strains in three clusters; grape-isolated strains formed one cluster, citrus-coffee strains formed another cluster, and a third cluster resulted from all other strains. Primer pairs designed for the 16S rRNA and gyrB genes were extensively searched in databases to verify their in silico specificity. Primer pairs were certified with 30 target and 36 nontarget pure cultures of microorganisms, confirming 100% specificity. A multiplex PCR protocol was developed and its sensitivity tested. Sequencing of PCR products confirmed the validity of the multiplex PCR. Xylella fastidiosa was detected in field-collected plants, disease vector insects, and nonsymptomatic but infected plants. Specific detection of X. fastidiosa may facilitate the understanding of its ecological significance and prevention of spread of the disease.
Figures


Similar articles
-
Assessment of the genetic diversity of Xylella fastidiosa isolated from citrus in Brazil by PCR-RFLP of the 16S rDNA and 16S-23S intergenic spacer and rep-PCR fingerprinting.Antonie Van Leeuwenhoek. 2001 Jan;79(1):53-9. doi: 10.1023/a:1010219811555. Antonie Van Leeuwenhoek. 2001. PMID: 11392484
-
A nested-PCR assay for detection of Xylella fastidiosa in citrus plants and sharpshooter leafhoppers.J Appl Microbiol. 2004;96(3):546-51. doi: 10.1111/j.1365-2672.2004.02176.x. J Appl Microbiol. 2004. PMID: 14962134
-
An evolutionary perspective of Pierce's disease of grapevine, citrus variegated chlorosis, and mulberry leaf scorch diseases.Curr Microbiol. 2002 Dec;45(6):423-8. doi: 10.1007/s00284-002-3785-7. Curr Microbiol. 2002. PMID: 12402083
-
The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology.Annu Rev Entomol. 2004;49:243-70. doi: 10.1146/annurev.ento.49.061802.123403. Annu Rev Entomol. 2004. PMID: 14651464 Review.
-
Paradigms: examples from the bacterium Xylella fastidiosa.Annu Rev Phytopathol. 2013;51:339-56. doi: 10.1146/annurev-phyto-082712-102325. Epub 2013 May 17. Annu Rev Phytopathol. 2013. PMID: 23682911 Review.
Cited by
-
Diversity Evaluation of Xylella fastidiosa from Infected Olive Trees in Apulia (Southern Italy).Plant Pathol J. 2016 Apr;32(2):102-11. doi: 10.5423/PPJ.OA.08.2015.0153. Epub 2016 Apr 1. Plant Pathol J. 2016. PMID: 27147930 Free PMC article.
-
Antimicrobial Peptides With Antibiofilm Activity Against Xylella fastidiosa.Front Microbiol. 2021 Nov 8;12:753874. doi: 10.3389/fmicb.2021.753874. eCollection 2021. Front Microbiol. 2021. PMID: 34819923 Free PMC article.
-
Xylella fastidiosa: Host Range and Advance in Molecular Identification Techniques.Front Plant Sci. 2017 Jun 8;8:944. doi: 10.3389/fpls.2017.00944. eCollection 2017. Front Plant Sci. 2017. PMID: 28642764 Free PMC article. Review.
-
Assessment of the Hyperspectral Data Analysis as a Tool to Diagnose Xylella fastidiosa in the Asymptomatic Leaves of Olive Plants.Plants (Basel). 2021 Apr 1;10(4):683. doi: 10.3390/plants10040683. Plants (Basel). 2021. PMID: 33916301 Free PMC article.
-
A multigene phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa.Appl Environ Microbiol. 2005 Jul;71(7):3832-9. doi: 10.1128/AEM.71.7.3832-3839.2005. Appl Environ Microbiol. 2005. PMID: 16000795 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

