Identification and tracing of Bifidobacterium species by use of enterobacterial repetitive intergenic consensus sequences
- PMID: 12839818
- PMCID: PMC165193
- DOI: 10.1128/AEM.69.7.4296-4301.2003
Identification and tracing of Bifidobacterium species by use of enterobacterial repetitive intergenic consensus sequences
Abstract
Eighty-nine Bifidobacterium strains from 26 species were identified and classified to the species level with an enterobacterial repetitive intergenic consensus (ERIC)-PCR approach. We demonstrated that ERIC-PCR is useful for a phylogenetic and taxonomical analysis but as well as for a species composition analysis of mixed bifidobacterial cultures isolated from dairy products and other environments.
Figures
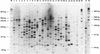

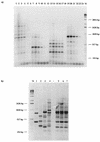
Similar articles
-
Identification of Bifidobacterium species using rep-PCR fingerprinting.Syst Appl Microbiol. 2003 Nov;26(4):557-63. doi: 10.1078/072320203770865864. Syst Appl Microbiol. 2003. PMID: 14666984
-
Intraspecies variability of Desulfovibrio desulfuricans strains determined by the genetic profiles.FEMS Microbiol Lett. 2003 Feb 14;219(1):69-74. doi: 10.1016/S0378-1097(02)01199-0. FEMS Microbiol Lett. 2003. PMID: 12594025
-
Typing of Haemophilus paragallinarum strains by using enterobacterial repetitive intergenic consensus-based polymerase chain reaction.Avian Dis. 2004 Dec;48(4):890-5. doi: 10.1637/7137. Avian Dis. 2004. PMID: 15666871
-
Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach.Appl Environ Microbiol. 2001 Jun;67(6):2760-5. doi: 10.1128/AEM.67.6.2760-2765.2001. Appl Environ Microbiol. 2001. PMID: 11375192 Free PMC article.
-
Genus- and species-specific PCR primers for the detection and identification of bifidobacteria.Curr Issues Intest Microbiol. 2003 Sep;4(2):61-9. Curr Issues Intest Microbiol. 2003. PMID: 14503690 Review.
Cited by
-
Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses.Appl Environ Microbiol. 2004 Oct;70(10):6197-209. doi: 10.1128/AEM.70.10.6197-6209.2004. Appl Environ Microbiol. 2004. PMID: 15466567 Free PMC article.
-
The Pork Food Chain as a Route of Transmission of Antimicrobial Resistant Escherichia coli: A Farm-to-Fork Perspective.Antibiotics (Basel). 2023 Feb 11;12(2):376. doi: 10.3390/antibiotics12020376. Antibiotics (Basel). 2023. PMID: 36830287 Free PMC article.
-
Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing.Appl Environ Microbiol. 2005 Jan;71(1):487-500. doi: 10.1128/AEM.71.1.487-500.2005. Appl Environ Microbiol. 2005. PMID: 15640225 Free PMC article.
-
Microbial characterization of probiotics--advisory report of the Working Group "8651 Probiotics" of the Belgian Superior Health Council (SHC).Mol Nutr Food Res. 2013 Aug;57(8):1479-504. doi: 10.1002/mnfr.201300065. Epub 2013 Jun 25. Mol Nutr Food Res. 2013. PMID: 23801655 Free PMC article.
-
Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis.Appl Environ Microbiol. 2003 Dec;69(12):7517-22. doi: 10.1128/AEM.69.12.7517-7522.2003. Appl Environ Microbiol. 2003. PMID: 14660406 Free PMC article.
References
-
- de Bruijn, F. 1992. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58:2180-2187. - PMC - PubMed
-
- Gillings, M., and M. Holley. 1997. Repetitive element PCR fingerprinting (rep-PCR) using enterobacterial repetitive intergenic consensus (ERIC) primers is not necessarily directed at ERIC elements. Lett. Appl. Microbiol. 25:17-21. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

