Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis
- PMID: 12839993
- PMCID: PMC165659
- DOI: 10.1093/emboj/cdg335
Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis
Abstract
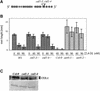
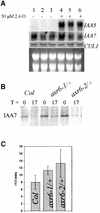
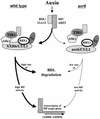
The AXR6 gene is required for auxin signaling in the Arabidopsis embryo and during postembryonic development. One of the effects of auxin is to stimulate degradation of the Aux/IAA auxin response proteins through the action of the ubiquitin protein ligase SCF(TIR1). Here we show that AXR6 encodes the SCF subunit CUL1. The axr6 mutations affect the ability of mutant CUL1 to assemble into stable SCF complexes resulting in reduced degradation of the SCF(TIR1) substrate AXR2/IAA7. In addition, we show that CUL1 is required for lateral organ initiation in the shoot apical meristem and the inflorescence meristem. These results indicate that the embryonic axr6 phenotype is related to a defect in SCF function and accumulation of Aux/IAA proteins such as BDL/IAA12. In addition, we show that CUL1 has a role in auxin response throughout the life cycle of the plant.
Figures





References
-
- Bennett S.R.M., Alvarez,J., Bossinger,G. and Smyth,D.R. (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J., 8, 505–520.
-
- Berleth T. and Jurgens,G. (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development, 118, 575–587.
-
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

