A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude
- PMID: 12839998
- PMCID: PMC165643
- DOI: 10.1093/emboj/cdg318
A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude
Abstract
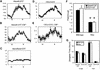
The transcription factor Clock (Clk) plays a critical role in animal circadian rhythms. Genetic studies defining its function have relied on two dominant negative alleles, one in Drosophila and one in mice. Here we describe a novel recessive allele of Drosophila Clock, Clk(ar). Homozygous Clk(ar) flies are viable and behaviorally arrhythmic. The Clk(ar) phenotype is caused by a splice site mutation that severely disrupts splicing and reduces Clk activity. Despite the behavioral arrhythmicity, molecular oscillations are still detectable in Clk(ar) flies. Transcription analysis indicates potent effects of Clk(ar) on levels and amplitude of transcriptional oscillations. Taken together with other data, we propose that Clk makes a major contribution to the strength and amplitude of circadian rhythms.
Figures

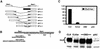



References
-
- Allada R., White,N.E., So,W.V., Hall,J.C. and Rosbash,M. (1998) A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell, 93, 791–804. - PubMed
-
- Allada R., Emery,P., Takahashi,J.S. and Rosbash,M. (2001) Stopping time: the genetics of fly and mouse circadian clocks. Annu. Rev. Neurosci., 24, 1091–1119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

