Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC
- PMID: 12840006
- PMCID: PMC165644
- DOI: 10.1093/emboj/cdg319
Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC
Abstract
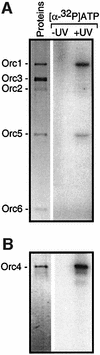
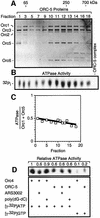
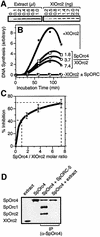
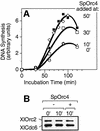
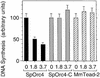
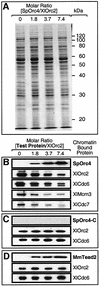
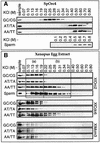
Budding yeast (Saccharomyces cerevisiae) origin recognition complex (ORC) requires ATP to bind specific DNA sequences, whereas fission yeast (Schizosaccharomyces pombe) ORC binds to specific, asymmetric A:T-rich sites within replication origins, independently of ATP, and frog (Xenopus laevis) ORC seems to bind DNA non-specifically. Here we show that despite these differences, ORCs are functionally conserved. Firstly, SpOrc1, SpOrc4 and SpOrc5, like those from other eukaryotes, bound ATP and exhibited ATPase activity, suggesting that ATP is required for pre-replication complex (pre-RC) assembly rather than origin specificity. Secondly, SpOrc4, which is solely responsible for binding SpORC to DNA, inhibited up to 70% of XlORC-dependent DNA replication in Xenopus egg extract by preventing XlORC from binding to chromatin and assembling pre-RCs. Chromatin-bound SpOrc4 was located at AT-rich sequences. XlORC in egg extract bound preferentially to asymmetric A:T-sequences in either bare DNA or in sperm chromatin, and it recruited XlCdc6 and XlMcm proteins to these sequences. These results reveal that XlORC initiates DNA replication preferentially at the same or similar sites to those targeted in S.pombe.
Figures
Similar articles
-
Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit.Mol Cell Biol. 2001 Dec;21(23):8095-103. doi: 10.1128/MCB.21.23.8095-8103.2001. Mol Cell Biol. 2001. PMID: 11689699 Free PMC article.
-
Role for a Xenopus Orc2-related protein in controlling DNA replication.Nature. 1996 Jan 25;379(6563):357-60. doi: 10.1038/379357a0. Nature. 1996. PMID: 8552193
-
ADP-binding to origin recognition complex of Saccharomyces cerevisiae.J Mol Biol. 2004 Jun 25;340(1):29-37. doi: 10.1016/j.jmb.2004.04.045. J Mol Biol. 2004. PMID: 15184020
-
Origins and complexes: the initiation of DNA replication.J Exp Bot. 2001 Feb;52(355):193-202. J Exp Bot. 2001. PMID: 11283163 Review.
-
DNA replication origins, ORC/DNA interaction, and assembly of pre-replication complex in eukaryotes.Acta Biochim Biophys Sin (Shanghai). 2010 Jul;42(7):433-9. doi: 10.1093/abbs/gmq048. Epub 2010 Jun 9. Acta Biochim Biophys Sin (Shanghai). 2010. PMID: 20705581 Review.
Cited by
-
TIF1 Represses rDNA replication initiation, but promotes normal S phase progression and chromosome transmission in Tetrahymena.Mol Biol Cell. 2005 Jun;16(6):2624-35. doi: 10.1091/mbc.e05-02-0107. Epub 2005 Mar 16. Mol Biol Cell. 2005. PMID: 15772155 Free PMC article.
-
The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo.EMBO J. 2006 Nov 15;25(22):5372-82. doi: 10.1038/sj.emboj.7601396. Epub 2006 Oct 26. EMBO J. 2006. PMID: 17066079 Free PMC article.
-
Regulation of DNA replication by chromatin structures: accessibility and recruitment.Chromosoma. 2011 Feb;120(1):39-46. doi: 10.1007/s00412-010-0287-4. Epub 2010 Aug 3. Chromosoma. 2011. PMID: 20680317 Review.
-
Genome-wide characterization of fission yeast DNA replication origins.EMBO J. 2006 Nov 1;25(21):5171-9. doi: 10.1038/sj.emboj.7601390. Epub 2006 Oct 19. EMBO J. 2006. PMID: 17053780 Free PMC article.
-
Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex's largest subunit (Orc1) from binding to chromatin during mitosis.Mol Cell Biol. 2004 Jul;24(13):5875-86. doi: 10.1128/MCB.24.13.5875-5886.2004. Mol Cell Biol. 2004. PMID: 15199143 Free PMC article.
References
-
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. - PubMed
-
- Alberts B. and Herrick,G. (1971) DNA cellulose chromatography. Methods Enzymol., 21, 198–217.
-
- Beall E.L., Manak,J.R., Zhou,S., Bell,M., Lipsick,J.S. and Botchan,M.R. (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature, 420, 833–837. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

