The balance of driving forces during genome evolution in prokaryotes
- PMID: 12840037
- PMCID: PMC403731
- DOI: 10.1101/gr.1092603
The balance of driving forces during genome evolution in prokaryotes
Abstract
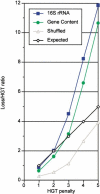
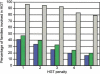
Genomes are shaped by evolutionary processes such as gene genesis, horizontal gene transfer (HGT), and gene loss. To quantify the relative contributions of these processes, we analyze the distribution of 12,762 protein families on a phylogenetic tree, derived from entire genomes of 41 Bacteria and 10 Archaea. We show that gene loss is the most important factor in shaping genome content, being up to three times more frequent than HGT, followed by gene genesis, which may contribute up to twice as many genes as HGT. We suggest that gene gain and gene loss in prokaryotes are balanced; thus, on average, prokaryotic genome size is kept constant. Despite the importance of HGT, our results indicate that the majority of protein families have only been transmitted by vertical inheritance. To test our method, we present a study of strain-specific genes of Helicobacter pylori, and demonstrate correct predictions of gene loss and HGT for at least 81% of validated cases. This approach indicates that it is possible to trace genome content history and quantify the factors that shape contemporary prokaryotic genomes.
Figures


References
-
- Alm, R.A., Ling, L.S., Moir, D.T., King, B.L., Brown, E.D., Doig, P.C., Smith, D.R., Noonan, B., Guild, B.C., deJonge, B.L., et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397: 176-180. - PubMed
-
- Andersson, J.O. 2000. Evolutionary genomics: Is Buchnera a bacterium or an organelle? Curr. Biol. 10: R866-R868. - PubMed
-
- Andersson, J.O. and Andersson, S.G. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9: 664-671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
