The spacing between adjacent binding sites in the family of repeats affects the functions of Epstein-Barr nuclear antigen 1 in transcription activation and stable plasmid maintenance
- PMID: 12842617
- PMCID: PMC2922029
- DOI: 10.1016/s0042-6822(03)00122-3
The spacing between adjacent binding sites in the family of repeats affects the functions of Epstein-Barr nuclear antigen 1 in transcription activation and stable plasmid maintenance
Abstract
Epstein-Barr virus (EBV) and the closely related Herpesvirus papio (HVP) are stably replicated as episomes in proliferating latently infected cells. Maintenance and partitioning of these viral plasmids requires a viral sequence in cis, termed the family of repeats (FR), that is bound by a viral protein, Epstein-Barr nuclear antigen 1 (EBNA1). Upon binding FR, EBNA1 maintains viral genomes in proliferating cells and activates transcription from viral promoters required for immortalization. FR from either virus encodes multiple binding sites for the viral maintenance protein, EBNA1, with the FR from the prototypic B95-8 strain of EBV containing 20 binding sites, and FR from HVP containing 8 binding sites. In addition to differences in the number of EBNA1-binding sites, adjacent binding sites in the EBV FR are typically separated by 14 base pairs (bp), but are separated by 10 bp in HVP. We tested whether the number of binding sites, as well as the distance between adjacent binding sites, affects the function of EBNA1 in transcription activation or plasmid maintenance. Our results indicate that EBNA1 activates transcription more efficiently when adjacent binding sites are separated by 10 bp, the spacing observed in HVP. In contrast, using two separate assays, we demonstrate that plasmid maintenance is greatly augmented when adjacent EBNA1-binding sites are separated by 14 bp, and therefore, presumably lie on the same face of the DNA double helix. These results provide indication that the functions of EBNA1 in transcription activation and plasmid maintenance are separable.
Figures


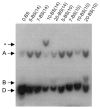
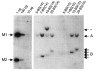
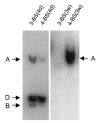
References
-
- Aiyar A, Sugden B. Fusions between Epstein–Barr viral nuclear antigen-1 of Epstein–Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J. Biol. Chem. 1998;273(49):33073–33081. - PubMed
-
- Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310(5974):207–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

