Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis
- PMID: 12843022
- PMCID: PMC165330
- DOI: 10.1128/JCM.41.7.2924-2931.2003
Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis
Abstract
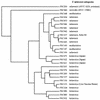
Francisella tularensis is a potent pathogen and a possible bioterrorism agent. Little is known, however, to explain the molecular basis for its virulence and the distinct differences in virulence found between the four recognized subspecies, F. tularensis subsp. tularensis, F. tularensis subsp. mediasiatica, F. tularensis subsp. holarctica, and F. tularensis subsp. novicida. We developed a DNA microarray based on 1,832 clones from a shotgun library used for sequencing of the highly virulent strain F. tularensis subsp. tularensis Schu S4. This allowed a genome-wide analysis of 27 strains representing all four subspecies. Overall, the microarray analysis confirmed a limited genetic variation within the species F. tularensis, and when the strains were compared, at most 3.7% of the probes showed differential hybridization. Cluster analysis of the hybridization data revealed that the causative agents of type A and type B tularemia, i.e., F. tularensis subsp. tularensis and F. tularensis subsp. holarctica, respectively, formed distinct clusters. Despite marked differences in their virulence and geographical origin, a high degree of genomic similarity between strains of F. tularensis subsp. tularensis and F. tularensis subsp. mediasiatica was apparent. Strains from Japan clustered separately, as did strains of F. tularensis subsp. novicida. Eight regions of difference (RD) 0.6 to 11.5 kb in size, altogether comprising 21 open reading frames, were identified that distinguished strains of the moderately virulent subspecies F. tularensis subsp. holarctica and the highly virulent subspecies F. tularensis subsp. tularensis. One of these regions, RD1, allowed for the first time the development of an F. tularensis-specific PCR assay that discriminates each of the four subspecies.
Figures

References
-
- Björkholm, B. M., J. L. Guruge, J. D. Oh, A. J. Syder, N. Salama, K. Guillemin, S. Falkow, C. Nilsson, P. G. Falk, L. Engstrand, and J. I. Gordon. 2002. Colonization of germ-free transgenic mice with genotyped Helicobacter pylori strains from a case-control study of gastric cancer reveals a correlation between host responses and HsdS components of type I restriction-modification systems. J. Biol. Chem. 277:34191-34197. - PubMed
-
- De Groote, M. A., T. Testerman, Y. Xu, G. Stauffer, and F. C. Fang. 1996. Homocysteine antagonism of nitric oxide-related cytostasis in Salmonella typhimurium. Science 272:414-417. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

