Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States
- PMID: 12843064
- PMCID: PMC165376
- DOI: 10.1128/JCM.41.7.3198-3205.2003
Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States
Abstract
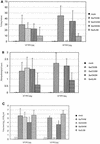
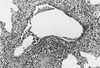
Swine influenza is an acute respiratory disease caused by type A influenza viruses. Before 1998, swine influenza virus isolates in the United States were mainly of the classical H1N1 lineage. Since then, phylogenetically distinct reassortant H3N2 viruses have been identified as respiratory pathogens in pigs on U.S. farms. The H3N2 viruses presently circulating in the U.S. swine population are triple reassortants containing avian-like (PA and PB2), swine-like (M, NP, and NS), and human-like (HA, NA, and PB1) gene segments. Recent sequence data show that the triple reassortants have acquired at least three distinct H3 molecules from human influenza viruses and thus form three distinct phylogenetic clusters (I to III). In this study we analyzed the antigenic and pathogenic properties of viruses belonging to each of these clusters. Hemagglutination inhibition and neutralization assays that used hyperimmune sera obtained from caesarian-derived, colostrum-deprived pigs revealed that H3N2 cluster I and cluster III viruses share common epitopes, whereas a cluster II virus showed only limited cross-reactivity. H3N2 viruses from each of the three clusters were able to induce clinical signs of disease and associated lesions upon intratracheal inoculation into seronegative pigs. There were, however, differences in the severity of lesions between individual strains even within one antigenic cluster. A correlation between the severity of disease and pig age was observed. These data highlight the increased diversity of swine influenza viruses in the United States and would indicate that surveillance should be intensified to determine the most suitable vaccine components.
Figures



References
-
- Choi, Y. K., S. M. Goyal, M. W. Farnham, and H. S. Joo. 2002. Phylogenetic analysis of H1N2 isolates of influenza A virus from pigs in the United States. Virus Res. 87:173-179. - PubMed
-
- Choi, Y. K., S. M. Goyal, S. W. Kang, M. W. Farnham, and H. S. Joo. 2002. Detection and subtyping of swine influenza H1N1, H1N2 and H3N2 viruses in clinical samples using two multiplex RT-PCR assays. J. Virol. Methods 102:53-59. - PubMed
-
- Easterday, B. C. 1980. Animals in the influenza world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 288:433-437. - PubMed
-
- Easterday, B. C., and V. S. Hinshaw. 1992. Swine influenza, p. 349-357. In A. D. Leman (ed.), Diseases of swine, 7th ed. Iowa State University Press, Ames.
-
- Heinen, P. P., A. P. van Nieuwstadt, E. A. de Boer-Luijtze, and A. T. Bianchi. 2001. Analysis of the quality of protection induced by a porcine influenza A vaccine to challenge with an H3N2 virus. Vet. Immunol. Immunopathol. 82:39-56. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

