Nucleotide sequence-based multitarget identification
- PMID: 12843076
- PMCID: PMC165273
- DOI: 10.1128/JCM.41.7.3284-3292.2003
Nucleotide sequence-based multitarget identification
Abstract
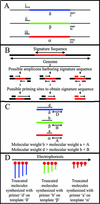
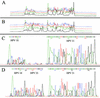
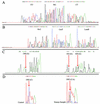
MULTIGEN technology (T. Vinayagamoorthy, U.S. patent 6,197,510, March 2001) is a modification of conventional sequencing technology that generates a single electropherogram consisting of short nucleotide sequences from a mixture of known DNA targets. The target sequences may be present on the same or different nucleic acid molecules. For example, when two DNA targets are sequenced, the first and second sequencing primers are annealed to their respective target sequences, and then a polymerase causes chain extension by the addition of new deoxyribose nucleotides. Since the electrophoretic separation depends on the relative molecular weights of the truncated molecules, the molecular weight of the second sequencing primer was specifically designed to be higher than the combined molecular weight of the first sequencing primer plus the molecular weight of the largest truncated molecule generated from the first target sequence. Thus, the series of truncated molecules produced by the second sequencing primer will have higher molecular weights than those produced by the first sequencing primer. Hence, the truncated molecules produced by these two sequencing primers can be effectively separated in a single lane by standard gel electrophoresis in a single electropherogram without any overlapping of the nucleotide sequences. By using sequencing primers with progressively higher molecular weights, multiple short DNA sequences from a variety of targets can be determined simultaneously. We describe here the basic concept of MULTIGEN technology and three applications: detection of sexually transmitted pathogens (Neisseria gonorrhoeae, Chlamydia trachomatis, and Ureaplasma urealyticum), detection of contaminants in meat samples (coliforms, fecal coliforms, and Escherichia coli O157:H7), and detection of single-nucleotide polymorphisms in the human N-acetyltransferase (NAT1) gene (S. Fronhoffs et al., Carcinogenesis 22:1405-1412, 2001).
Figures
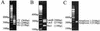
Similar articles
-
[Detection of Neisseria gonorrhoeae(NG), Chlamydia trachomatis(CT) and Ureaplasma urealyticum(UU) by multiple primer PCR].Wei Sheng Wu Xue Bao. 2000 Apr;40(2):221-3. Wei Sheng Wu Xue Bao. 2000. PMID: 12548950 Chinese.
-
Fabrication and optimization of the multiplex PCR-based oligonucleotide microarray for detection of Neisseria gonorrhoeae, Chlamydia trachomatis and Ureaplasma urealyticum.J Microbiol Methods. 2005 Aug;62(2):245-56. doi: 10.1016/j.mimet.2005.02.017. J Microbiol Methods. 2005. PMID: 15893829
-
Comparison of rRNA-based and DNA-based nucleic acid amplifications for detection of Chlamydia trachomatis, Neisseria gonorrhoeae, and Ureaplasma urealyticum in urogenital swabs.BMC Infect Dis. 2018 Dec 12;18(1):651. doi: 10.1186/s12879-018-3580-0. BMC Infect Dis. 2018. PMID: 30541468 Free PMC article.
-
Molecular tests for detection of the sexually-transmitted pathogens Neisseria gonorrhoeae and Chlamydia trachomatis.Med Health R I. 2006 Jun;89(6):202-4. Med Health R I. 2006. PMID: 16875007 Review. No abstract available.
-
Cobas® 4800: a fully automated system for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae.Expert Rev Mol Diagn. 2013 Mar;13(2):131-40. doi: 10.1586/erm.12.141. Expert Rev Mol Diagn. 2013. PMID: 23477553 Review.
Cited by
-
High prevalence of oncogenic HPV-16 in cervical smears of asymptomatic women of eastern Uttar Pradesh, India: a population-based study.J Biosci. 2012 Mar;37(1):63-72. doi: 10.1007/s12038-012-9181-y. J Biosci. 2012. PMID: 22357204
-
Detection of BRAF mutations from solid tumors using Tumorplex™ technology.MethodsX. 2015 Jun 12;2:316-22. doi: 10.1016/j.mex.2015.06.002. eCollection 2015. MethodsX. 2015. PMID: 26258049 Free PMC article.
-
Identification of the severe acute respiratory syndrome coronavirus by simultaneous multigene DNA sequencing.J Clin Microbiol. 2004 Jul;42(7):3291-4. doi: 10.1128/JCM.42.7.3291-3294.2004. J Clin Microbiol. 2004. PMID: 15243096 Free PMC article.
-
Detection of human papillomavirus DNA in cervical samples: analysis of the new PGMY-PCR compared to the hybrid capture II and MY-PCR assays and a two-step nested PCR assay.J Clin Microbiol. 2004 Aug;42(8):3861-4. doi: 10.1128/JCM.42.8.3861-3864.2004. J Clin Microbiol. 2004. PMID: 15297550 Free PMC article.
-
Molecular typing of West Nile Virus, Dengue, and St. Louis encephalitis using multiplex sequencing.J Mol Diagn. 2005 May;7(2):152-9. doi: 10.1016/S1525-1578(10)60541-7. J Mol Diagn. 2005. PMID: 15858138 Free PMC article.
References
-
- Afshari, C. A., E. F. Nuwaysir, and J. C. Barrett. 1999. Application of complementary DNA microarray technology to carcinogen identification, toxicology, and drug safety evaluation. Cancer Res. 59:4759-4760. - PubMed
-
- Aono, T., K. Kondo, H. Miyoshi, K. Tanaka-Taya, M. Kondo, Y. Osugi, J. Hara, S. Okada, and K. Yamanishi. 1998. Monitoring of human cytomegalovirus infections in pediatric bone marrow transplant recipients by nucleic acid sequence-based amplification. J. Infect. Dis. 178:1244-1249. - PubMed
-
- Bonora, S., M. C. Gutierrez, G. Di-Perri, F. Brunello, B. Allegranzi, M. Ligozzi, R. Fontana, E. Concia, and V. Vincent. 1999. Comparative evaluation of ligation-mediated PCR and spoligotyping as screening methods for genotyping of Mycobacterium tuberculios strains. J. Clin. Microbiol. 37:3118-3123. - PMC - PubMed
-
- Church, G. M., and S. Kieffer-Higgins. 1988. Multiplex DNA sequencing. Science 240:185-188. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

