Rapid detection of Candida albicans in clinical blood samples by using a TaqMan-based PCR assay
- PMID: 12843077
- PMCID: PMC165319
- DOI: 10.1128/JCM.41.7.3293-3298.2003
Rapid detection of Candida albicans in clinical blood samples by using a TaqMan-based PCR assay
Abstract
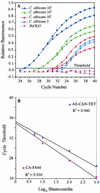
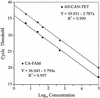
We describe a rapid and reproducible PCR assay for quantitation of the Candida albicans ribosomal DNA (rDNA) in clinical blood samples based on the TaqMan principle (Applied Biosystems), in which a signal is generated by cleavage of a template-specific probe during amplification. We used two fluorogenic probes based on universal, fungus-specific primers, one for the detection of C. albicans species DNA and one for the detection of all Candida genus DNA. C. albicans blastoconidia mixed with whole blood in a titration experiment yielded a linear PCR signal over a range of 3 orders of magnitude. The TaqMan-based PCR assay for C. albicans exhibited a low limit of detection (5 CFU/ml of blood) and an excellent reproducibility (96 to 99%). While the C. albicans species-specific probe had 100% specificity for C. albicans, all Candida genus-specific probes cross-reacted with other organisms likely to coinfect patients with C. albicans infections. On the basis of these data, we determined the C. albicans loads with a species-specific probe from 122 blood samples from 61 hematology or oncology patients with clinically proven or suspected systemic Candida infections. Eleven positive samples exhibited a wide range of C. albicans loads, extending from 5 to 100,475 CFU/ml of blood. The sensitivity and specificity of the present assay were 100 and 97%, respectively, compared with the results of blood culture. These data indicate that the TaqMan-based PCR assay for quantitation of C. albicans with a species-specific probe provides an attractive alternative for the identification and quantitation of C. albicans rDNA in pure cultures and blood samples.
Figures
Similar articles
-
Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis.J Clin Microbiol. 2000 Aug;38(8):3016-21. doi: 10.1128/JCM.38.8.3016-3021.2000. J Clin Microbiol. 2000. PMID: 10921970 Free PMC article.
-
Improving molecular detection of Candida DNA in whole blood: comparison of seven fungal DNA extraction protocols using real-time PCR.J Med Microbiol. 2008 Mar;57(Pt 3):296-303. doi: 10.1099/jmm.0.47617-0. J Med Microbiol. 2008. PMID: 18287291
-
Seminested PCR for diagnosis of candidemia: comparison with culture, antigen detection, and biochemical methods for species identification.J Clin Microbiol. 2002 Jul;40(7):2483-9. doi: 10.1128/JCM.40.7.2483-2489.2002. J Clin Microbiol. 2002. PMID: 12089267 Free PMC article.
-
Molecular diagnosis and epidemiology of fungal infections.Med Mycol. 1998;36 Suppl 1:249-57. Med Mycol. 1998. PMID: 9988514 Review.
-
Advances in Candida detection platforms for clinical and point-of-care applications.Crit Rev Biotechnol. 2017 Jun;37(4):441-458. doi: 10.3109/07388551.2016.1167667. Epub 2016 Apr 19. Crit Rev Biotechnol. 2017. PMID: 27093473 Free PMC article. Review.
Cited by
-
PCR-detectable Candida DNA exists a short period in the blood of systemic candidiasis murine model.Open Life Sci. 2020 Sep 6;15(1):677-682. doi: 10.1515/biol-2020-0075. eCollection 2020. Open Life Sci. 2020. PMID: 33817256 Free PMC article.
-
Molecular Identification of Candida albicans in Endodontic Retreatment Cases by SYBR Green I Real-time Polymerase Chain Reaction and its Association with Endodontic Symptoms.J Dent (Shiraz). 2023 Dec 1;24(4):429-437. doi: 10.30476/dentjods.2023.91706.1786. eCollection 2023 Dec. J Dent (Shiraz). 2023. PMID: 38149236 Free PMC article.
-
Development and validation of a quantitative PCR assay using multiplexed hydrolysis probes for detection and quantification of Theileria orientalis isolates and differentiation of clinically relevant subtypes.J Clin Microbiol. 2015 Mar;53(3):941-50. doi: 10.1128/JCM.03387-14. Epub 2015 Jan 14. J Clin Microbiol. 2015. PMID: 25588653 Free PMC article.
-
An Italian consensus for invasive candidiasis management (ITALIC).Infection. 2014 Apr;42(2):263-79. doi: 10.1007/s15010-013-0558-0. Epub 2013 Nov 25. Infection. 2014. PMID: 24272916 Review.
-
Large scale multiplex PCR improves pathogen detection by DNA microarrays.BMC Microbiol. 2009 Jan 3;9:1. doi: 10.1186/1471-2180-9-1. BMC Microbiol. 2009. PMID: 19121223 Free PMC article.
References
-
- Beck-Sagué, C., and W. R. Jarvis, et al. 1993. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980-1990. J. Infect. Dis. 167:1247-1251. - PubMed
-
- Borst, A., M. A. Leverstein-Van Hall, J. Verhoef, and A. C. Fluit. 2001. Detection of Candida spp. in blood cultures using nucleic acid sequence-based amplification (NASBA). Diagn. Microbiol. Infect. Dis. 39:155-160. - PubMed
-
- Bowman, J. C., G. K. Abruzzo, J. W. Anderson, A. M. Flattery, C. J. Gill, V. B. Pikounis, D. M. Schmatz, P. A. Liberator, and C. M., Douglas. 2001. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob. Agents Chemother. 45:3474-3481. - PMC - PubMed
-
- Chen, Y. C., J. D. Eisner, M. M. Katar, S. L. Rassoulian-Barrett, K. LaFe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeast using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

