Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus
- PMID: 12847288
- PMCID: PMC166420
- DOI: 10.1073/pnas.1332765100
Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus
Abstract
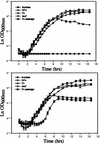
Lactobacillus acidophilus is a probiotic organism that displays the ability to use prebiotic compounds such as fructooligosaccharides (FOS), which stimulate the growth of beneficial commensals in the gastrointestinal tract. However, little is known about the mechanisms and genes involved in FOS utilization by Lactobacillus species. Analysis of the L. acidophilus NCFM genome revealed an msm locus composed of a transcriptional regulator of the LacI family, a four-component ATP-binding cassette (ABC) transport system, a fructosidase, and a sucrose phosphorylase. Transcriptional analysis of this operon demonstrated that gene expression was induced by sucrose and FOS but not by glucose or fructose, suggesting some specificity for nonreadily fermentable sugars. Additionally, expression was repressed by glucose but not by fructose, suggesting catabolite repression via two cre-like sequences identified in the promoter-operator region. Insertional inactivation of the genes encoding the ABC transporter substrate-binding protein and the fructosidase reduced the ability of the mutants to grow on FOS. Comparative analysis of gene architecture within this cluster revealed a high degree of synteny with operons in Streptococcus mutans and Streptococcus pneumoniae. However, the association between a fructosidase and an ABC transporter is unusual and may be specific to L. acidophilus. This is a description of a previously undescribed gene locus involved in transport and catabolism of FOS compounds, which can promote competition of beneficial microorganisms in the human gastrointestinal tract.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

