Sequential signaling through Notch1 and erbB receptors mediates radial glia differentiation
- PMID: 12853432
- PMCID: PMC6740346
- DOI: 10.1523/JNEUROSCI.23-14-06132.2003
Sequential signaling through Notch1 and erbB receptors mediates radial glia differentiation
Abstract
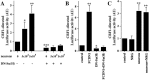
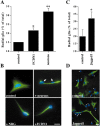
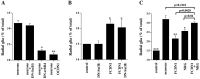
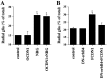
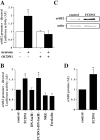
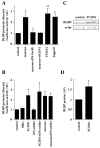
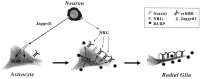
Radial glia cells both generate neurons and physically guide nascent neurons to their target destination in the cortex, and as such they are essential for CNS development. It has been proposed that in the developing cerebellum, neuronal contact induces radial glia formation, however, the mechanisms involved in this process are not well understood. Here we demonstrate that neuronal induction of radial glia formation is the result of sequential signaling through Notch1 and erbB receptors. First, Notch1 activation by neuronal contact induces the glial expression of the brain lipid binding protein (BLBP) and erbB2 genes. Interestingly, two different signaling pathways mediate these effects of Notch1 on transcription, BLBP expression being dependent on Su(H), whereas erbB2 is regulated by a yet unidentified Notch1 pathway. The subsequent increase in erbB2 receptor expression makes the glia more responsive to neuronal NRG, which then induces the morphological transformation into radial glia. Thus, these results unveil some of the mechanisms underlying radial glia formation.
Figures

References
-
- Alvarez-Buylla A, Theelen M, Nottebohm F ( 1990) Proliferation “hot spots” in adult avian ventricular zone reveal radial cell division. Neuron 5: 101–109. - PubMed
-
- Anton ES, Marchionni MA, Lee KF, Rakic P ( 1997) Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development 124: 3501–3510. s - PubMed
-
- Canoll PD, Musacchio JM, Hardy R, Reynolds R, Marchionni MA, Salzer JL ( 1996) GGF/neuregulin is a neuronal signal that promotes the proliferation and survival and inhibits the differentiation of oligodendrocyte progenitors. Neuron 17: 229–243. - PubMed
-
- Chanas-Sacre G, Rogister B, Moonen G, Leprince P ( 2000) Radial glia phenotype: origin, regulation, and transdifferentiation. J Neurosci Res 61: 357–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
