Direct real-time molecular scale visualisation of the degradation of condensed DNA complexes exposed to DNase I
- PMID: 12853616
- PMCID: PMC165977
- DOI: 10.1093/nar/gkg462
Direct real-time molecular scale visualisation of the degradation of condensed DNA complexes exposed to DNase I
Abstract
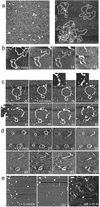
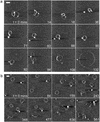
The need to protect DNA from in vivo degradation is one of the basic tenets of therapeutic gene delivery and a standard test for any proposed delivery vector. The currently employed in vitro tests, however, presently provide no direct link between the molecular structure of the vector complexes and their success in this role, thus hindering the rational design of successful gene delivery agents. Here we apply atomic force microscopy (AFM) in liquid to visualise at the molecular scale and in real time, the effect of DNase I on generation 4 polyamidoamine dendrimers (G4) complexed with DNA. These complexes are revealed to be dynamic in nature showing a degree of mobility, in some cases revealing the addition and loss of dendrimers to individual complexes. The formation of the G4-DNA complexes is observed to provide a degree of protection to the DNA. This protection is related to the structural morphology of the formed complex, which is itself shown to be dependent on the dendrimer loading and the time allowed for complex formation.
Figures

Similar articles
-
Structural study of DNA condensation induced by novel phosphorylcholine-based copolymers for gene delivery and relevance to DNA protection.Langmuir. 2005 Apr 12;21(8):3591-8. doi: 10.1021/la047480i. Langmuir. 2005. PMID: 15807606
-
The interaction of plasmid DNA with polyamidoamine dendrimers: mechanism of complex formation and analysis of alterations induced in nuclease sensitivity and transcriptional activity of the complexed DNA.Biochim Biophys Acta. 1997 Aug 7;1353(2):180-90. doi: 10.1016/s0167-4781(97)00069-9. Biochim Biophys Acta. 1997. PMID: 9294012
-
Bundling and aggregation of DNA by cationic dendrimers.Biomacromolecules. 2011 Feb 14;12(2):511-7. doi: 10.1021/bm1013102. Epub 2010 Dec 30. Biomacromolecules. 2011. PMID: 21192723
-
Multi-armed poly(L-glutamic acid)-graft-polypropyleneinime as effective and serum resistant gene delivery vectors.Int J Pharm. 2014 Apr 25;465(1-2):444-54. doi: 10.1016/j.ijpharm.2014.02.041. Epub 2014 Feb 25. Int J Pharm. 2014. PMID: 24576809
-
[Progress in the studies of DNA-protein interactions by atomic force microscopy].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2007 Oct;24(5):1172-6. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2007. PMID: 18027720 Review. Chinese.
Cited by
-
Visual analysis of concerted cleavage by type IIF restriction enzyme SfiI in subsecond time region.Biophys J. 2011 Dec 21;101(12):2992-8. doi: 10.1016/j.bpj.2011.09.064. Epub 2011 Dec 20. Biophys J. 2011. PMID: 22208198 Free PMC article.
-
Improving the Enzymatic Stability and the Pharmacokinetics of Oligonucleotides via DNA-Backboned Bottlebrush Polymers.Nano Lett. 2018 Nov 14;18(11):7378-7382. doi: 10.1021/acs.nanolett.8b03662. Epub 2018 Nov 2. Nano Lett. 2018. PMID: 30376347 Free PMC article.
-
Gene therapy of benign gynecological diseases.Adv Drug Deliv Rev. 2009 Aug 10;61(10):822-35. doi: 10.1016/j.addr.2009.04.023. Epub 2009 May 13. Adv Drug Deliv Rev. 2009. PMID: 19446586 Free PMC article. Review.
-
Combinatorial evaluation of cations, pH-sensitive and hydrophobic moieties for polymeric vector design.Mol Ther. 2009 Mar;17(3):480-90. doi: 10.1038/mt.2008.293. Epub 2009 Jan 13. Mol Ther. 2009. PMID: 19142180 Free PMC article.
-
Visualizing the attack of RNase enzymes on dendriplexes and naked RNA using atomic force microscopy.PLoS One. 2013 Apr 18;8(4):e61710. doi: 10.1371/journal.pone.0061710. Print 2013. PLoS One. 2013. PMID: 23637890 Free PMC article.
References
-
- Hill I.R.C., Garnett,M.C., Bignotti,F. and Davis,S.S. (2001) Determination of protection from serum nuclease activity by DNA–polyelectrolyte complexes using an electrophoretic method. Anal. Biochem., 291, 62–68. - PubMed
-
- Bielinska A.U., Kukowska-Latallo,J.F. and Baker,J.R.,Jr (1997) The interaction of plasmid DNA with polyamidoamine dendrimers: mechanism of complex formation and analysis of alteration induced in nuclease sensitivity and transcriptional activity of the complexed DNA. Biochim. Biophys. Acta, 1353, 180–190. - PubMed
-
- Tang M.X., Redemann,C.T. and Szoka F.C. (1996) In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjugate Chem., 7, 703–714. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

