Spatial organization of topoisomerase I-mediated DNA cleavage induced by camptothecin-oligonucleotide conjugates
- PMID: 12853620
- PMCID: PMC165972
- DOI: 10.1093/nar/gkg457
Spatial organization of topoisomerase I-mediated DNA cleavage induced by camptothecin-oligonucleotide conjugates
Abstract
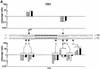
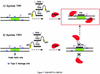
Triple helix-forming oligonucleotides covalently linked to topoisomerase I inhibitors, in particular the antitumor agent camptothecin, trigger topoisomerase I-mediated DNA cleavage selectively in the proximity of the binding site of the oligonucleotide vector. In the present study, we have performed a systematic analysis of the DNA cleavage efficiency as a function of the positioning of the camptothecin derivative, either on the 3' or the 5' side of the triplex, and the location of the cleavage site. A previously identified cleavage site was inserted at different positions within two triplex site-containing 59 bp duplexes. Sequence-specific DNA cleavage by topoisomerase I occurs only with triplex conjugates bearing the inhibitor at the 3'-end of the oligonucleotide and on the oligopyrimidine strand of the duplex. The lack of targeted cleavage on the 5' side is attributed to the structural differences of the 3' and 5' duplex-triplex DNA junctions. The changes induced in the double helix by the triple-helical structure interfere with the action of the enzyme according to a preferred spatial organization. Camptothecin conjugates of oligonucleotides provide efficient tools to probe the organization of the topoisomerase I-DNA complex and will be useful to understand the functioning of topoisomerase I in living cells.
Figures
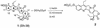



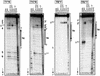

References
-
- Champoux J.J. (2001) DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem., 70, 369–413. - PubMed
-
- Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. - PubMed
-
- Pommier Y., Pourquier,P., Fan,Y. and Strumberg,D. (1998) Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta, 1400, 83–105. - PubMed
-
- Redinbo M.R., Stewart,L., Kuhn,P., Champoux,J.J. and Hol,W.G. (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science, 279, 1504–1513. - PubMed
-
- Stewart L., Redinbo,M.R., Qiu,X., Hol,W.G.J. and Champoux,J.J. (1998) A model for the mechanism of human topoisomerase I. Science, 279, 1534–1541. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

