The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity
- PMID: 12853623
- PMCID: PMC165952
- DOI: 10.1093/nar/gkg437
The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity
Abstract
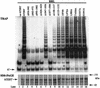
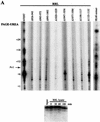
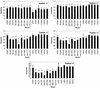
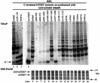
The catalytic subunit of telomerase (TERT) contains conserved reverse transcriptase-like motifs but N- and C-terminal regions unique to telomerases. Despite weak sequence conservation, the C terminus of TERTs from various organisms has been implicated in telomerase-specific functions, including telomerase activity, functional multimerization with other TERT molecules, enzyme processivity and telomere length maintenance. We studied hTERT proteins containing small C-terminal deletions or substitutions to identify and characterize hTERT domains mediating telomerase activity, hTERT multimerization and processivity. Using sequence alignment of five vertebrate TERTs and Arabidopsis thaliana TERT, we identified blocks of highly conserved amino acids that were required for human telomerase activity and functional hTERT complementation. We adapted the non-PCR-based telomerase elongation assay to characterize telomerase expressed and reconstituted in the in vitro transcription/translation rabbit reticulocyte lysate system. Using this assay, we found that the hTERT C terminus, like the C terminus of Saccharomyces cerevisiae TERT, contributes to successive nucleotide addition within a single 6-base telomeric repeat (type I processivity). Certain mutations in the hTERT C terminus also reduced the repetitive addition of multiple telomeric repeats (type II processivity). Our results suggest a functionally conserved role for the TERT C terminus in telomerase enzyme processivity.
Figures




References
-
- Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. - PubMed
-
- Harrington L. and Robinson,M.O. (2002) Telomere dysfunction: multiple paths to the same end. Oncogene, 21, 592–597. - PubMed
-
- Blackburn E. (1999) Telomerase. In Gestland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 609–635.
-
- Feng J., Funk,W.D., Wang,S.-S., Weinrich,S.L., Avilion,A.A., Chiu,C.-P., Adams,R.R., Chang,E., Allsopp,R.C., Yu,J., Le,S., West,M.D., Harley,C.B., Andrews,W.H., Greider,C.W. and Villeponteau,B. (1995) The RNA component of human telomerase. Science, 269, 1236–1241. - PubMed
-
- Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

