CADLIVE for constructing a large-scale biochemical network based on a simulation-directed notation and its application to yeast cell cycle
- PMID: 12853624
- PMCID: PMC165976
- DOI: 10.1093/nar/gkg461
CADLIVE for constructing a large-scale biochemical network based on a simulation-directed notation and its application to yeast cell cycle
Abstract
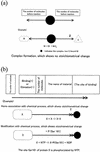
The further understanding of the mechanisms of gene regulatory networks requires comprehensive tools for both the representation of complicated signal transduction pathways and the in silico identification of genomic signals that govern the regulation of gene expression. Consequently, sophisticated notation must be developed to represent the signal transduction pathways in a form that can be readily processed by both computers and humans. We propose the regulator-reaction equations combined with detailed attributes including the associated cellular component, molecular function, and biological process and present the simulation-directed graphical notation that is derived from modification of Kohn's method. We have developed the software suite, CADLIVE (Computer-Aided Design of LIVing systEms), which features a graphical user interface (GUI) to edit large-scale maps of complicated signal transduction pathways using a conventional XML-based representation. The regulator-reaction equations represent not only mechanistic reactions, but also semantic models containing ambiguous and incomplete processes. In order to demonstrate the feasibility of CADLIVE, we constructed a detailed map of the budding yeast cell cycle, which consists of 184 molecules and 152 reactions, in a really compact space. CADLIVE enables one to look at the whole view of a large-scale map, to integrate postgenomic data into the map, and to computationally simulate the signal transduction pathways, which greatly facilitates exploring novel or unexpected interactions.
Figures






Similar articles
-
Extended CADLIVE: a novel graphical notation for design of biochemical network maps and computational pathway analysis.Nucleic Acids Res. 2007;35(20):e134. doi: 10.1093/nar/gkm769. Epub 2007 Oct 16. Nucleic Acids Res. 2007. PMID: 17940089 Free PMC article.
-
CADLIVE dynamic simulator: direct link of biochemical networks to dynamic models.Genome Res. 2005 Apr;15(4):590-600. doi: 10.1101/gr.3463705. Genome Res. 2005. PMID: 15805500 Free PMC article.
-
Statistical and integrative approach for constructing biological network maps.Genome Inform. 2004;15(2):161-70. Genome Inform. 2004. PMID: 15706502
-
A comprehensive molecular interaction map of the budding yeast cell cycle.Mol Syst Biol. 2010 Sep 21;6:415. doi: 10.1038/msb.2010.73. Mol Syst Biol. 2010. PMID: 20865008 Free PMC article. Review.
-
Cellular quiescence in budding yeast.Yeast. 2021 Jan;38(1):12-29. doi: 10.1002/yea.3545. Epub 2021 Jan 25. Yeast. 2021. PMID: 33350503 Free PMC article. Review.
Cited by
-
Graphical Modeling Meets Systems Pharmacology.Gene Regul Syst Bio. 2017 Mar 10;11:1177625017691937. doi: 10.1177/1177625017691937. eCollection 2017. Gene Regul Syst Bio. 2017. PMID: 28469411 Free PMC article.
-
An efficient grid layout algorithm for biological networks utilizing various biological attributes.BMC Bioinformatics. 2007 Mar 6;8:76. doi: 10.1186/1471-2105-8-76. BMC Bioinformatics. 2007. PMID: 17338825 Free PMC article.
-
Molecular interaction maps of bioregulatory networks: a general rubric for systems biology.Mol Biol Cell. 2006 Jan;17(1):1-13. doi: 10.1091/mbc.e05-09-0824. Epub 2005 Nov 2. Mol Biol Cell. 2006. PMID: 16267266 Free PMC article.
-
Improvement of the memory function of a mutual repression network in a stochastic environment by negative autoregulation.BMC Bioinformatics. 2019 Dec 27;20(1):734. doi: 10.1186/s12859-019-3315-2. BMC Bioinformatics. 2019. PMID: 31881978 Free PMC article.
-
A new grid- and modularity-based layout algorithm for complex biological networks.PLoS One. 2019 Aug 29;14(8):e0221620. doi: 10.1371/journal.pone.0221620. eCollection 2019. PLoS One. 2019. PMID: 31465473 Free PMC article.
References
-
- Abbott A. (1999) Alliance of US labs plans to build map of cell signalling pathways. Nature, 402, 219–220. - PubMed
-
- Kitano H. (2002) Systems biology: a brief overview. Science, 295, 1662–1664. - PubMed
-
- Pirson I., Fortemaison,N., Jacobs,C., Dremier,S., Dumont,J.E. and Maenhaut,C. (2000) The visual display of regulatory information and networks. Trends Cell Biol., 10, 404–408. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

