Hidden messages in the nef gene of human immunodeficiency virus type 1 suggest a novel RNA secondary structure
- PMID: 12853637
- PMCID: PMC165969
- DOI: 10.1093/nar/gkg454
Hidden messages in the nef gene of human immunodeficiency virus type 1 suggest a novel RNA secondary structure
Abstract
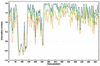
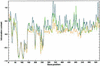
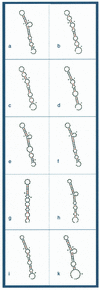
The coexistence of multiple codes in the genome of human immunodeficiency virus type 1 (HIV-1) was analyzed. We explored factors constraining the variability of the virus genome primarily in relation to conserved RNA secondary structures overlapping coding sequences, and used a simple combination of algorithms for RNA secondary structure prediction based on the nearest-neighbor thermodynamic rules and a statistical approach. In our previous study, we applied this combination to a non- redundant data set of env nucleotide sequences, confirmed the conservative secondary structure of the rev-responsive element (RRE) and found a new RNA structure in the first conserved (C1) region of the env gene. In this study, we analyzed the variability of putative RNA secondary structures inside the nef gene of HIV-1 by applying these algorithms to a non-redundant data set of 104 nef sequences retrieved from the Los Alamos HIV database, and predicted the existence of a novel functional RNA secondary structure in the beta3/beta4 regions of nef. The predicted RNA fold in the beta3/beta4 region of nef appears in two forms with different loop sizes. The loop of the first fold consists of seven nucleotides (positions 494-500), with consensus UCAAGCU appearing in 79% of sequences. The other has a five-base loop (positions 495-499) with consensus CAAGC. The difference in size between these two loops may reflect the difference between respective counterparts in the hairpin recognition. This may also have an adaptive biological significance.
Figures

Similar articles
-
RNA secondary structure and squence conservation in C1 region of human immunodeficiency virus type 1 env gene.AIDS Res Hum Retroviruses. 2002 Aug 10;18(12):867-78. doi: 10.1089/08892220260190353. AIDS Res Hum Retroviruses. 2002. PMID: 12201910
-
Persistence of attenuated HIV-1 rev alleles in an epidemiologically linked cohort of long-term survivors infected with nef-deleted virus.Retrovirology. 2007 Jul 1;4:43. doi: 10.1186/1742-4690-4-43. Retrovirology. 2007. PMID: 17601342 Free PMC article.
-
A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression.J Virol. 2004 Jun;78(12):6517-26. doi: 10.1128/JVI.78.12.6517-6526.2004. J Virol. 2004. PMID: 15163745 Free PMC article.
-
Regulation of expression of human immunodeficiency virus.New Biol. 1990 Jan;2(1):20-31. New Biol. 1990. PMID: 2078551 Review.
-
Points to ponder on the function of Nef.Res Virol. 1992 Jan-Feb;143(1):47-9. doi: 10.1016/s0923-2516(06)80078-6. Res Virol. 1992. PMID: 1565855 Review. No abstract available.
Cited by
-
Overlapping messages and survivability.J Mol Evol. 2004 Oct;59(4):520-7. doi: 10.1007/s00239-004-2644-5. J Mol Evol. 2004. PMID: 15638463
-
On the role of four small hairpins in the HIV-1 RNA genome.RNA Biol. 2013 Apr;10(4):540-52. doi: 10.4161/rna.24133. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535706 Free PMC article.
-
Evidence of a novel RNA secondary structure in the coding region of HIV-1 pol gene.RNA. 2008 Dec;14(12):2478-88. doi: 10.1261/rna.1252608. Epub 2008 Oct 30. RNA. 2008. PMID: 18974280 Free PMC article.
-
Changes in the Plasticity of HIV-1 Nef RNA during the Evolution of the North American Epidemic.PLoS One. 2016 Sep 29;11(9):e0163688. doi: 10.1371/journal.pone.0163688. eCollection 2016. PLoS One. 2016. PMID: 27685447 Free PMC article.
-
Cassandra retrotransposons carry independently transcribed 5S RNA.Proc Natl Acad Sci U S A. 2008 Apr 15;105(15):5833-8. doi: 10.1073/pnas.0709698105. Epub 2008 Apr 11. Proc Natl Acad Sci U S A. 2008. PMID: 18408163 Free PMC article.
References
-
- Trifonov E.N. (1989) The multiple codes of nucleotide sequences. Bull. Math. Biol., 51, 417–432. - PubMed
-
- Trifonov E.N. (1990) Making sense of the human genome. In Sarma,R.H. and Sarma,M.H. (eds), Structure and Methods. Adenine Press, Albany, NY, Vol. 1, pp. 69–77.
-
- Trifonov E.N. (1996) Interfering contexts of regulatory sequence elements. CABIOS, 12, 423–429. - PubMed
-
- Dayton E., Powell,D. and Dayton,A. (1989) Functional analysis of CAR, the target sequence for the Rev protein of HIV-1. Science, 246, 1625–1629. - PubMed
-
- Malim M., Hauber,J., Le,S.-Y., Maizel,J. and Cullen,B. (1989) The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature, 338, 254–257. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

