A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons
- PMID: 12857733
- PMCID: PMC2635476
- DOI: 10.1074/jbc.M305580200
A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons
Abstract
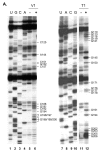
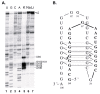
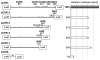
The 484-nucleotide (nt) alternatively translated region (ATR) of the human fibroblast growth factor 2 (FGF-2) mRNA contains four CUG and one AUG translation initiation codons. Although the 5'-end proximal CUG codon is initiated by a cap-dependent translation process, the other four initiation codons are initiated by a mechanism of internal entry of ribosomes. We undertook here a detailed analysis of the cis-acting elements defining the FGF-2 internal ribosome entry site (IRES). A thorough deletion analysis study within the 5'-ATR led us to define a 176-nt region as being necessary and sufficient for IRES function at four codons present in a downstream 308-nt RNA segment. Unexpectedly, a single IRES module is therefore responsible for translation initiation at four distantly localized codons. The determination of the FGF-2 5'-ATR RNA secondary structure by enzymatic and chemical probing experiments showed that the FGF-2 IRES contained two stem-loop regions and a G quartet motif that constitute novel structural determinants of IRES function.
Figures

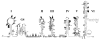



References
-
- Bikfalvi A, Klein S, Pintucci G, Rifkin DB. Endocr Rev. 1997;18:26–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

