Localization of nonspecific lipid transfer proteins correlate with programmed cell death responses during endosperm degradation in Euphorbia lagascae seedlings
- PMID: 12857807
- PMCID: PMC167065
- DOI: 10.1104/pp.103.020875
Localization of nonspecific lipid transfer proteins correlate with programmed cell death responses during endosperm degradation in Euphorbia lagascae seedlings
Abstract
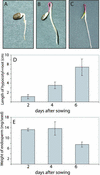
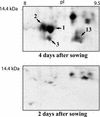
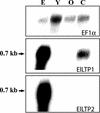
When the storage materials have been depleted, the endosperm cells undergo programmed cell death. Very little is known about how the components of the dying cells are recycled and used by the growing seedling. To learn more about endosperm degradation and nutrient recycling, we isolated soluble proteins from the endosperm of Euphorbia lagascae seedlings collected 2, 4, and 6 d after sowing. The protein extracts were subjected to two-dimensional gel electrophoresis. Proteins that increased in amount in the endosperm with time were selected for further analysis with mass spectrometry. We successfully identified 17 proteins, which became more abundant by time during germination. Among these proteins were three E. lagascae lipid transfer proteins (ElLTPs), ElLTP1, ElLTP2, and ElLTP3. Detailed expressional studies were performed on ElLTP1 and ElLTP2. ElLTP1 transcripts were detected in endosperm and cotyledons, whereas ElLTP2 transcripts were only detected in endosperm. Western blots confirmed that ElLTP1 and ElLTP2 accumulate during germination. Immunolocalization experiments showed that ElLTP1 was present in the vessels of the developing cotyledons, and also in the alloplastic space in the endosperm. ElLTP2 formed a concentration gradient in the endosperm, with higher amounts in the inner regions close to the cotyledons, and lesser amounts in the outer regions of the endosperm. On the basis of these data, we propose that ElLTP1 and ElLTP2 are involved in recycling of endosperm lipids, or that they act as protease inhibitors protecting the growing cotyledons from proteases released during programmed cell death.
Figures




Similar articles
-
Characterization of germination-specific lipid transfer proteins from Euphorbia lagascae.Planta. 2002 May;215(1):41-50. doi: 10.1007/s00425-001-0717-x. Epub 2002 Feb 2. Planta. 2002. PMID: 12012240
-
Characterization of SCP-2 from Euphorbia lagascae reveals that a single Leu/Met exchange enhances sterol transfer activity.FEBS J. 2006 Dec;273(24):5641-55. doi: 10.1111/j.1742-4658.2006.05553.x. FEBS J. 2006. PMID: 17212780
-
A germination-specific epoxide hydrolase from Euphorbia lagascae.Planta. 2003 Jan;216(3):403-12. doi: 10.1007/s00425-002-0876-4. Epub 2002 Oct 2. Planta. 2003. PMID: 12520331
-
On the role of a Lipid-Transfer Protein. Arabidopsis ltp3 mutant is compromised in germination and seedling growth.Plant Signal Behav. 2015;10(12):e1105417. doi: 10.1080/15592324.2015.1105417. Plant Signal Behav. 2015. PMID: 26479260 Free PMC article.
-
The Maltase Involved in Starch Metabolism in Barley Endosperm Is Encoded by a Single Gene.PLoS One. 2016 Mar 24;11(3):e0151642. doi: 10.1371/journal.pone.0151642. eCollection 2016. PLoS One. 2016. PMID: 27011041 Free PMC article.
Cited by
-
XSP10 and SlSAMT, Fusarium wilt disease responsive genes of tomato (Solanum lycopersicum L.) express tissue specifically and interact with each other at cytoplasm in vivo.Physiol Mol Biol Plants. 2021 Jul;27(7):1559-1575. doi: 10.1007/s12298-021-01025-y. Epub 2021 Jun 28. Physiol Mol Biol Plants. 2021. PMID: 34366597 Free PMC article.
-
Responses to telomere erosion in plants.PLoS One. 2014 Jan 21;9(1):e86220. doi: 10.1371/journal.pone.0086220. eCollection 2014. PLoS One. 2014. PMID: 24465970 Free PMC article.
-
Molecular Control of Oil Metabolism in the Endosperm of Seeds.Int J Mol Sci. 2021 Feb 5;22(4):1621. doi: 10.3390/ijms22041621. Int J Mol Sci. 2021. PMID: 33562710 Free PMC article. Review.
-
Insight into the molecular evolution of non-specific lipid transfer proteins via comparative analysis between rice and sorghum.DNA Res. 2012 Apr;19(2):179-94. doi: 10.1093/dnares/dss003. Epub 2012 Feb 24. DNA Res. 2012. PMID: 22368182 Free PMC article.
-
A combined transcriptomic and proteomic analysis of chrysanthemum provides new insights into petal senescence.Planta. 2021 Dec 16;255(1):22. doi: 10.1007/s00425-021-03808-9. Planta. 2021. PMID: 34918180
References
-
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol 133: 1251–1263 - PMC - PubMed
-
- Beevers H (1980) The role of the glyoxylate cycle, In PK Stumpf, ed, The Biochemistry of Plants, Vol 4. Academic Press, New York, pp 117–130
-
- Bergman P, Edqvist J, Farbos I, Glimelius K (2000) Male-sterile tobacco displays abnormal mitochondrial atp1 transcript accumulation and reduced floral ATP/ADP ratio. Plant Mol Biol 42: 531–544 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

