Mitochondrial phosphatidylserine decarboxylase from higher plants. Functional complementation in yeast, localization in plants, and overexpression in Arabidopsis
- PMID: 12857846
- PMCID: PMC167104
- DOI: 10.1104/pp.103.023242
Mitochondrial phosphatidylserine decarboxylase from higher plants. Functional complementation in yeast, localization in plants, and overexpression in Arabidopsis
Abstract
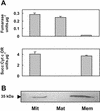
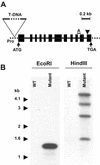
Plants are known to synthesize ethanolamine (Etn) moieties by decarboxylation of free serine (Ser), but there is also some evidence for phosphatidyl-Ser (Ptd-Ser) decarboxylation. Database searches identified diverse plant cDNAs and an Arabidopsis gene encoding 50-kD proteins homologous to yeast (Saccharomyces cerevisiae) and mammalian mitochondrial Ptd-Ser decarboxylases (PSDs). Like the latter, the plant proteins have putative mitochondrial targeting and inner membrane sorting sequences and contain near the C terminus a Glycine-Serine-Threonine motif corresponding to the site of proteolysis and catalytic pyruvoyl residue formation. A truncated tomato (Lycopersicon esculentum) cDNA lacking the targeting sequence and a chimeric construct in which the targeting and sorting sequences were replaced by those from yeast PSD1 both complemented the Etn requirement of a yeast psd1 psd2 mutant, and PSD activity was detected in the mitochondria of the complemented cells. Immunoblot analysis of potato (Solanum tuberosum) mitochondria demonstrated that PSD is located in mitochondrial membranes, and mRNA analysis in Arabidopsis showed that the mitochondrial PSD gene is expressed at low levels throughout the plant. An Arabidopsis knockup mutant grew normally but had 6- to 13-fold more mitochondrial PSD mRNA and 9-fold more mitochondrial PSD activity. Total membrane PSD activity was, however, unchanged in the mutant, showing mitochondrial activity to be a minor part of the total. These results establish that plants can synthesize Etn moieties via a phospholipid pathway and have both mitochondrial and extramitochondrial PSDs. They also indicate that mitochondrial PSD is an important housekeeping enzyme whose expression is strongly regulated at the transcriptional level.
Figures



Similar articles
-
Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme.J Biol Chem. 2001 Sep 21;276(38):35523-9. doi: 10.1074/jbc.M106038200. Epub 2001 Jul 18. J Biol Chem. 2001. PMID: 11461929
-
Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria.Plant Physiol. 2007 Jun;144(2):904-14. doi: 10.1104/pp.107.095414. Epub 2007 Apr 20. Plant Physiol. 2007. PMID: 17449644 Free PMC article.
-
Phosphatidylserine decarboxylase.Biochim Biophys Acta. 1997 Sep 4;1348(1-2):236-44. doi: 10.1016/s0005-2760(97)00101-x. Biochim Biophys Acta. 1997. PMID: 9370338 Review.
-
Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiáe. Cloning and mapping of the gene, heterologous expression, and creation of the null allele.J Biol Chem. 1995 Mar 17;270(11):6071-80. doi: 10.1074/jbc.270.11.6071. J Biol Chem. 1995. PMID: 7890740
-
Phosphatidylserine decarboxylases, key enzymes of lipid metabolism.IUBMB Life. 2009 Feb;61(2):151-62. doi: 10.1002/iub.159. IUBMB Life. 2009. PMID: 19165886 Review.
Cited by
-
Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1.J Biol Chem. 2012 Oct 26;287(44):36744-55. doi: 10.1074/jbc.M112.398107. Epub 2012 Sep 13. J Biol Chem. 2012. PMID: 22984266 Free PMC article.
-
Sinorhizobium meliloti mutants deficient in phosphatidylserine decarboxylase accumulate phosphatidylserine and are strongly affected during symbiosis with alfalfa.J Bacteriol. 2008 Oct;190(20):6846-56. doi: 10.1128/JB.00610-08. Epub 2008 Aug 15. J Bacteriol. 2008. PMID: 18708506 Free PMC article.
-
Acyl-lipid metabolism.Arabidopsis Book. 2013;11:e0161. doi: 10.1199/tab.0161. Epub 2013 Jan 29. Arabidopsis Book. 2013. PMID: 23505340 Free PMC article.
-
Lipid transport between the endoplasmic reticulum and mitochondria.Cold Spring Harb Perspect Biol. 2013 Jun 1;5(6):a013235. doi: 10.1101/cshperspect.a013235. Cold Spring Harb Perspect Biol. 2013. PMID: 23732475 Free PMC article. Review.
-
Glycerolipid synthesis and lipid trafficking in plant mitochondria.FEBS J. 2017 Feb;284(3):376-390. doi: 10.1111/febs.13812. Epub 2016 Aug 1. FEBS J. 2017. PMID: 27406373 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

