Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress
- PMID: 12857861
- PMCID: PMC165673
- DOI: 10.1091/mbc.e02-11-0740
Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress
Abstract
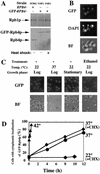
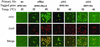
Changes in gene expression represent a major mechanism by which cells respond to stress. We and other investigators have previously shown that the yeast RNA polymerase II subunit Rpb4p is required for transcription under various stress conditions, but not under optimal growth conditions. Here we show that, in addition to its role in transcription, Rpb4p is also required for mRNA export, but only when cells are exposed to stress conditions. The roles of Rpb4p in transcription and in mRNA export can be uncoupled genetically by specific mutations in Rpb4p. Both functions of Rpb4p are required to maintain cell viability during stress. We propose that Rpb4p participates in the cellular responses to stress at the interface of the transcription and the export machineries.
Figures





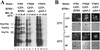
References
-
- Amberg, D.C., Goldstein, A.L., and Cole, C.N. (1992). Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6, 1173–1189. - PubMed
-
- Buratowski, S. (2000). Snapshots of RNA polymerase II transcription initiation. Curr. Opin. Cell Biol. 12, 320–325. - PubMed
-
- Choder, M. (1991). A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 5, 2315–2326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

