Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes
- PMID: 12857888
- PMCID: PMC165229
- DOI: 10.1128/jvi.77.15.8196-8206.2003
Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes
Abstract
Retroviruses copy their RNA genome into a DNA molecule, but little is known of the structure of the complex mediating reverse transcription in vivo. We used confocal and electron microscopy to study the structure of human immunodeficiency virus type 1 (HIV-1) intracellular reverse transcription complexes (RTCs). Cytoplasmic extracts were prepared 3, 4, and 16 h after acute infection by Dounce homogenization in hypotonic buffer. RTCs were purified by velocity sedimentation, followed by density fractionation in linear sucrose gradients and dialysis in a large pore cellulose membrane. RTCs had a sedimentation velocity of approximately 350 S and a density of 1.34 g/ml and were active in an endogenous reverse transcription assay. Double labeling of nucleic acids and viral proteins allowed specific visualization of RTCs by confocal microscopy. Electron microscopy revealed that RTCs are large nucleoprotein structures of variable shape consisting of packed filaments ca. 6 nm thick. Integrase and Vpr are associated with discrete regions of the 6-nm filaments. The nucleic acids within the RTC are coated by small proteins distinct from nucleocapsid and are partially protected from nuclease digestion.
Figures

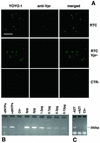
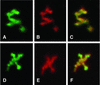

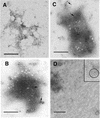
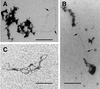

References
-
- Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediated the integration of retroviral DNA. Genes Dev. 3:469-478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

