Long-term latent murine Gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo
- PMID: 12857900
- PMCID: PMC165249
- DOI: 10.1128/jvi.77.15.8310-8321.2003
Long-term latent murine Gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo
Abstract
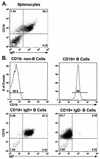
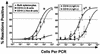
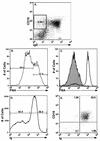
Murine gammaherpesvirus 68 (gammaHV68; also known as MHV-68) can establish a latent infection in both inbred and outbred strains of mice and, as such, provides a tractable small-animal model to address mechanisms and cell types involved in the establishment and maintenance of chronic gammaherpesvirus infection. Latency can be established at multiple anatomic sites, including the spleen and peritoneum; however, the contribution of distinct cell types to the maintenance of latency within these reservoirs remains poorly characterized. B cells are the major hematopoietic cell type harboring latent gammaHV68. We have analyzed various splenic B-cell subsets at early, intermediate, and late times postinfection and determined the frequency of cells either (i) capable of spontaneously reactivating latent gammaHV68 or (ii) harboring latent viral genome. These analyses demonstrated that latency is established in a variety of cell populations but that long-term latency (6 months postinfection) in the spleen after intranasal inoculation predominantly occurs in B cells. Furthermore, at late times postinfection latent gammaHV68 is largely confined to the surface immunoglobulin D-negative subset of B cells.
Figures



References
-
- Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. - PubMed
-
- Babcock, G. J., D. Hochberg, and A. D. Thorley-Lawson. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13:497-506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

