Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations
- PMID: 12857956
- PMCID: PMC166396
- DOI: 10.1073/pnas.1133470100
Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations
Abstract
Many areas of biomedical research depend on the analysis of uncommon variations in individual genes or transcripts. Here we describe a method that can quantify such variation at a scale and ease heretofore unattainable. Each DNA molecule in a collection of such molecules is converted into a single magnetic particle to which thousands of copies of DNA identical in sequence to the original are bound. This population of beads then corresponds to a one-to-one representation of the starting DNA molecules. Variation within the original population of DNA molecules can then be simply assessed by counting fluorescently labeled particles via flow cytometry. This approach is called BEAMing on the basis of four of its principal components (beads, emulsion, amplification, and magnetics). Millions of individual DNA molecules can be assessed in this fashion with standard laboratory equipment. Moreover, specific variants can be isolated by flow sorting and used for further experimentation. BEAMing can be used for the identification and quantification of rare mutations as well as to study variations in gene sequences or transcripts in specific populations or tissues.
Figures


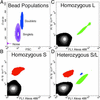
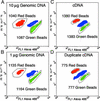
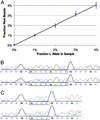
References
-
- Collins, F. S., Patrinos, A., Jordan, E., Chakravarti, A., Gesteland, R. & Walters, L. (1998) Science 282, 682–689. - PubMed
-
- Vogelstein, B. & Kinzler, K. W. (2002) The Genetic Basis of Human Cancer (McGraw–Hill, Toronto).
-
- Sidransky, D., Von Eschenbach, A., Tsai, Y. C., Jones, P., Summerhayes, I., Marshall, F., Paul, M., Green, P., Hamilton, S. R., Frost, P., et al. (1991) Science 252, 706–709. - PubMed
-
- Ahlquist, D. A. & Shuber, A. P. (2002) Clin. Chim. Acta 315, 157–168. - PubMed
-
- Sidransky, D. (2002) Nat. Rev. Cancer 2, 210–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

