QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling
- PMID: 12860968
- PMCID: PMC2172803
- DOI: 10.1083/jcb.200212083
QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling
Abstract
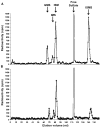
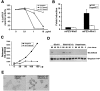
The 6-O sulfation states of cell surface heparan sulfate proteoglycans (HSPGs) are dynamically regulated to control the growth and specification of embryonic progenitor lineages. However, mechanisms for regulation of HSPG sulfation have been unknown. Here, we report on the biochemical and Wnt signaling activities of QSulf1, a novel cell surface sulfatase. Biochemical studies establish that QSulf1 is a heparan sulfate (HS) 6-O endosulfatase with preference, in particular, toward trisulfated IdoA2S-GlcNS6S disaccharide units within HS chains. In cells, QSulf1 can function cell autonomously to remodel the sulfation of cell surface HS and promote Wnt signaling when localized either on the cell surface or in the Golgi apparatus. QSulf1 6-O desulfation reduces XWnt binding to heparin and HS chains of Glypican1, whereas heparin binds with high affinity to XWnt8 and inhibits Wnt signaling. CHO cells mutant for HS biosynthesis are defective in Wnt-dependent Frizzled receptor activation, establishing that HS is required for Frizzled receptor function. Together, these findings suggest a two-state "catch or present" model for QSulf1 regulation of Wnt signaling in which QSulf1 removes 6-O sulfates from HS chains to promote the formation of low affinity HS-Wnt complexes that can functionally interact with Frizzled receptors to initiate Wnt signal transduction.
Figures




References
-
- Avnur, Z., and B. Geiger. 1984. Immunocytochemical localization of native chondroitin-sulfate in tissues and cultured cells using specific monoclonal antibody. Cell. 38:811–822. - PubMed
-
- Baeg, G.H., X. Lin, N. Khare, S. Baumgartner, and N. Perrimon. 2001. Heparan sulfate proteoglycans are critical for the organization of the extracellular distribution of Wingless. Development. 128:87–94. - PubMed
-
- Bai, X., K.J. Bame, H. Habuchi, K. Kimata, and J.D. Esko. 1997. Turnover of heparan sulfate depends on 2-O sulfation of uronic acids. J. Biol. Chem. 272:23172–23179. - PubMed
-
- Bai, X., G. Wei, A. Sinha, and J.D. Esko. 1999. Chinese hamster ovary cell mutants defective in glycosaminoglycan assembly and glucuronosyltransferase I. J. Biol. Chem. 274:13017–13024. - PubMed
-
- Bernfield, M., M. Gotte, P.W. Park, O. Reizes, M.L. Fitzgerald, J. Lincecum, and M. Zako. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68:729–777. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

