Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8
- PMID: 12861006
- PMCID: PMC165714
- DOI: 10.1128/MCB.23.15.5198-5207.2003
Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8
Abstract
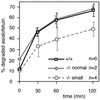
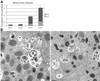
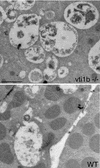
SNARE proteins participate in recognition and fusion of membranes. A SNARE complex consisting of vti1b, syntaxin 8, syntaxin 7, and endobrevin/VAMP-8 which is required for fusion of late endosomes in vitro has been identified recently. Here, we generated mice deficient in vti1b to study the function of this protein in vivo. vti1b-deficient mice had reduced amounts of syntaxin 8 due to degradation of the syntaxin 8 protein, while the amounts of syntaxin 7 and endobrevin did not change. These data indicate that vti1b is specifically required for the stability of a single SNARE partner. vti1b-deficient mice were viable and fertile. Most vti1b-deficient mice were indistinguishable from wild-type mice and did not display defects in transport to the lysosome. However, 20% of the vti1b-deficient mice were smaller. Lysosomal degradation of an endocytosed protein was slightly delayed in hepatocytes derived from these mice. Multivesicular bodies and autophagic vacuoles accumulated in hepatocytes of some smaller vti1b-deficient mice. This suggests that other SNAREs can compensate for the reduction in syntaxin 8 and for the loss of vti1b in most mice even though vti1b shows only 30% amino acid identity with its closest relative.
Figures

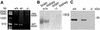





References
-
- Advani, R. J., H. R. Bae, J. B. Bock, D. S. Chao, Y. C. Doung, R. Prekeris, J. S. Yoo, and R. H. Scheller. 1998. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J. Biol. Chem. 273:10317-10324. - PubMed
-
- Antonin, W., D. Fasshauer, S. Becker, R. Jahn, and T. Schneider. 2002. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9:107-111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
