IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis
- PMID: 12867459
- PMCID: PMC165757
- DOI: 10.1128/JB.185.15.4499-4507.2003
IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis
Abstract
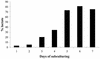
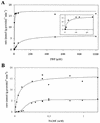
Lactococcus lactis NZ9010 in which the las operon-encoded ldh gene was replaced with an erythromycin resistance gene cassette displayed a stable phenotype when grown under aerobic conditions, and its main end products of fermentation under these conditions were acetate and acetoin. However, under anaerobic conditions, the growth of these cells was strongly retarded while the main end products of fermentation were acetate and ethanol. Upon prolonged subculturing of this strain under anaerobic conditions, both the growth rate and the ability to produce lactate were recovered after a variable number of generations. This recovery was shown to be due to the transcriptional activation of a silent ldhB gene coding for an Ldh protein (LdhB) with kinetic parameters different from those of the native las operon-encoded Ldh protein. Nevertheless, cells producing LdhB produced mainly lactate as the end product of fermentation. The mechanism underlying the ldhB gene activation was primarily studied in a single-colony isolate of the recovered culture, designated L. lactis NZ9015. Integration of IS981 in the upstream region of ldhB was responsible for transcription activation of the ldhB gene by generating an IS981-derived -35 promoter region at the correct spacing with a natively present -10 region. Subsequently, analysis of 10 independently isolated lactate-producing derivatives of L. lactis NZ9010 confirmed that the ldhB gene is transcribed in all of them. Moreover, characterization of the upstream region of the ldhB gene in these derivatives indicated that site-specific and directional IS981 insertion represents the predominant mechanism of the observed recovery of the ability to produce lactate.
Figures




References
-
- Andersen, H. W., M. B. Pedersen, K. Hammer, and P. R. Jensen. 2001. Lactate dehydrogenase has no control on lactate production but has a strong negative control on formate production in Lactococcus lactis. Eur. J. Biochem. 268:6379-6389. - PubMed
-
- Bhowmik, T., and J. L. Steele. 1994. Cloning, characterization and insertional inactivation of the Lactobacillus helveticus D(−) lactate dehydrogenase gene. Appl. Microbiol. Biotechnol. 41:432-439. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

