Characterization of transcriptional regulation of Shewanella frigidimarina Fe(III)-induced flavocytochrome c reveals a novel iron-responsive gene regulation system
- PMID: 12867466
- PMCID: PMC165765
- DOI: 10.1128/JB.185.15.4564-4571.2003
Characterization of transcriptional regulation of Shewanella frigidimarina Fe(III)-induced flavocytochrome c reveals a novel iron-responsive gene regulation system
Abstract
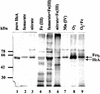
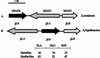
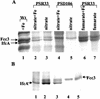
The bacterium Shewanella frigidimarina can grow anaerobically by utilizing Fe(III) as a respiratory electron acceptor. This results in the synthesis of a number of periplasmic c-type cytochromes, which are absent when the organism is grown in the absence of added Fe(III). One cytochrome, IfcA, is synthesized when Fe(III) is present as the sole respiratory electron acceptor or when it is present in combination with oxygen, fumarate, or nitrate. The ifcA gene was thus selected for a study of iron-responsive gene regulation of respiratory proteins in S. frigidimarina. The monocistronic ifcA gene clusters with two other monocistronic genes, ifcO, encoding a putative outer membrane porin, and ifcR, encoding a putative transcriptional regulator of the LysR superfamily. Analysis of transcription of all three genes under a range of growth conditions in the wild type and an ifcR insertion mutant and analysis of a strain that constitutively expresses ifcR revealed that iron regulation is exerted at the level of ifcR transcription. In the presence of Fe(III) IfcR is synthesized and acts positively to regulate expression of ifcO and ifcA. Control of Fe(III) respiration by this novel regulatory system differs markedly from Fur-mediated regulation of iron assimilation, in which Fur serves as an Fe(II)-activated repressor.
Figures




Similar articles
-
Characterization of a flavocytochrome that is induced during the anaerobic respiration of Fe3+ by Shewanella frigidimarina NCIMB400.Biochem J. 1999 Sep 1;342 ( Pt 2)(Pt 2):439-48. Biochem J. 1999. PMID: 10455032 Free PMC article.
-
The outer membrane protein Omp35 affects the reduction of Fe(III), nitrate, and fumarate by Shewanella oneidensis MR-1.BMC Microbiol. 2004 Jun 22;4:23. doi: 10.1186/1471-2180-4-23. BMC Microbiol. 2004. PMID: 15212692 Free PMC article.
-
Transcriptional and proteomic analysis of a ferric uptake regulator (fur) mutant of Shewanella oneidensis: possible involvement of fur in energy metabolism, transcriptional regulation, and oxidative stress.Appl Environ Microbiol. 2002 Feb;68(2):881-92. doi: 10.1128/AEM.68.2.881-892.2002. Appl Environ Microbiol. 2002. PMID: 11823232 Free PMC article.
-
The membrane-bound tetrahaem c-type cytochrome CymA interacts directly with the soluble fumarate reductase in Shewanella.Biochem Soc Trans. 2002 Aug;30(4):658-62. doi: 10.1042/bst0300658. Biochem Soc Trans. 2002. PMID: 12196158 Review.
-
Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria.Front Cell Infect Microbiol. 2013 Oct 2;3:59. doi: 10.3389/fcimb.2013.00059. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 24106689 Free PMC article. Review.
Cited by
-
Assessment of data processing to improve reliability of microarray experiments using genomic DNA reference.BMC Genomics. 2008 Sep 16;9 Suppl 2(Suppl 2):S5. doi: 10.1186/1471-2164-9-S2-S5. BMC Genomics. 2008. PMID: 18831796 Free PMC article.
-
Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors.J Bacteriol. 2005 Oct;187(20):7138-45. doi: 10.1128/JB.187.20.7138-7145.2005. J Bacteriol. 2005. PMID: 16199584 Free PMC article.
-
Shewanella oneidensis MR-1 sensory box protein involved in aerobic and anoxic growth.Appl Environ Microbiol. 2011 Jul;77(13):4647-56. doi: 10.1128/AEM.03003-10. Epub 2011 May 20. Appl Environ Microbiol. 2011. PMID: 21602393 Free PMC article.
-
Expression of hurP, a gene encoding a prospective site 2 protease, is essential for heme-dependent induction of bhuR in Bordetella bronchiseptica.J Bacteriol. 2007 Sep;189(17):6266-75. doi: 10.1128/JB.00629-07. Epub 2007 Jun 22. J Bacteriol. 2007. PMID: 17586630 Free PMC article.
-
Proteomic studies of an Antarctic cold-adapted bacterium, Shewanella livingstonensis Ac10, for global identification of cold-inducible proteins.Extremophiles. 2007 Nov;11(6):819-26. doi: 10.1007/s00792-007-0098-6. Epub 2007 Jul 7. Extremophiles. 2007. PMID: 17618403
References
-
- Bamford, V., P. S. Dobbin, D. J. Richardson, and A. M. Hemmings. 1999. Open conformation of a flavocytochrome c3 fumarate reductase. Nat. Struct. Biol. 6:1104-1107. - PubMed
-
- Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

