The cell death regulator GRIM-19 is an inhibitor of signal transducer and activator of transcription 3
- PMID: 12867595
- PMCID: PMC170920
- DOI: 10.1073/pnas.1633516100
The cell death regulator GRIM-19 is an inhibitor of signal transducer and activator of transcription 3
Abstract
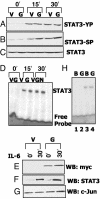
GRIM-19 (gene associated with retinoid-IFN-induced mortality 19), isolated as a cell death activator in a genetic screen used to define mechanisms involved in IFN-beta- and retinoic acid-induced cell death, codes for a approximately 16-kDa protein that induces apoptosis in a number of cell lines. Antisense ablation of GRIM-19 caused resistance to cell death induced by IFN plus retinoic acid and conferred a growth advantage to cells. To understand the molecular bases for its cell death regulatory activity, we used a yeast two-hybrid screen and identified that the transcription factor STAT3 (signal transducer and activator of transcription 3) binds to GRIM-19. GRIM-19 inhibits transcription driven by activation of STAT3, but not STAT1. It neither inhibits the ligand-induced activation of STAT3 nor blocks its ability to bind to DNA. Mutational analysis indicates that the transactivation domain of STAT3, especially residue S727, is required for GRIM-19 binding. Because GRIM-19 does not bind significantly to other STATs, our studies identify a specific inhibitor of STAT3. Because constitutively active STAT3 up-regulates antiapoptotic genes to promote tumor survival, its inhibition by GRIM-19 also demonstrates an antioncogenic effect exerted by biological therapeutics.
Figures




References
-
- Kimchi, A. (1998) Biochim. Biophys. Acta 1377, F13–F33. - PubMed
-
- Kalvakolanu, D. V. (2000) Histol. Histopathol. 15, 523–537. - PubMed
-
- Ikeda, H., Old, L. J. & Schreiber, R. D. (2002) Cytokine Growth Factor Rev. 13, 95–109. - PubMed
-
- Willman, C. L., Sever, C. E., Pallavicini, M. G., Harada, H., Tanaka, N., Slovak, M. L., Yamamoto, H., Harada, K., Meeker, T. C., List, A. F. & Taniguchi, T. (1993) Science 259, 968–971. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

