Biopipe: a flexible framework for protocol-based bioinformatics analysis
- PMID: 12869579
- PMCID: PMC403782
- DOI: 10.1101/gr.1363103
Biopipe: a flexible framework for protocol-based bioinformatics analysis
Abstract
We identify several challenges facing bioinformatics analysis today. Firstly, to fulfill the promise of comparative studies, bioinformatics analysis will need to accommodate different sources of data residing in a federation of databases that, in turn, come in different formats and modes of accessibility. Secondly, the tsunami of data to be handled will require robust systems that enable bioinformatics analysis to be carried out in a parallel fashion. Thirdly, the ever-evolving state of bioinformatics presents new algorithms and paradigms in conducting analysis. This means that any bioinformatics framework must be flexible and generic enough to accommodate such changes. In addition, we identify the need for introducing an explicit protocol-based approach to bioinformatics analysis that will lend rigorousness to the analysis. This makes it easier for experimentation and replication of results by external parties. Biopipe is designed in an effort to meet these goals. It aims to allow researchers to focus on protocol design. At the same time, it is designed to work over a compute farm and thus provides high-throughput performance. A common exchange format that encapsulates the entire protocol in terms of the analysis modules, parameters, and data versions has been developed to provide a powerful way in which to distribute and reproduce results. This will enable researchers to discuss and interpret the data better as the once implicit assumptions are now explicitly defined within the Biopipe framework.
Figures


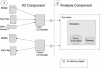





References
-
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al.2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. - PubMed
-
- Burge, C. and Karlin, S.1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. - PubMed
-
- Chicurel, M.2002. Bioinformatics: Bringing it all together. Nature 419: 751-755. - PubMed
-
- Davidson, S.B., Overton, C., and Buneman, P. 1995. Challenges in integrating biological data sources. J. Comput. Biol. 2: 557-572. - PubMed
WEB SITE REFERENCES
-
- ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/; CLUSTALW.
-
- http://blast.wustl.edu; BLAST.
-
- http://cvs.bioperl.org/cgi-bin/viewcvs/viewcvs.cgi/bioperl-pipeline/?cvs...; live CVS of the Biopipe source code.
-
- http://evolution.genetics.washington.edu/phylip.html; PHYLIP Package.
-
- http://genome.ucsc.edu; genome database.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
