SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response
- PMID: 12869585
- PMCID: PMC196237
- DOI: 10.1101/gad.1107803
SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response
Abstract
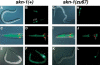
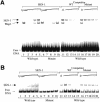
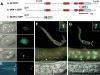
During the earliest stages of Caenorhabditis elegans embryogenesis, the transcription factor SKN-1 initiates development of the digestive system and other mesendodermal tissues. Postembryonic SKN-1 functions have not been elucidated. SKN-1 binds to DNA through a unique mechanism, but is distantly related to basic leucine-zipper proteins that orchestrate the major oxidative stress response in vertebrates and yeast. Here we show that despite its distinct mode of target gene recognition, SKN-1 functions similarly to resist oxidative stress in C. elegans. During postembryonic stages, SKN-1 regulates a key Phase II detoxification gene through constitutive and stress-inducible mechanisms in the ASI chemosensory neurons and intestine, respectively. SKN-1 is present in ASI nuclei under normal conditions, and accumulates in intestinal nuclei in response to oxidative stress. skn-1 mutants are sensitive to oxidative stress and have shortened lifespans. SKN-1 represents a connection between developmental specification of the digestive system and one of its most basic functions, resistance to oxidative and xenobiotic stress. This oxidative stress response thus appears to be both widely conserved and ancient, suggesting that the mesendodermal specification role of SKN-1 was predated by its function in these detoxification mechanisms.
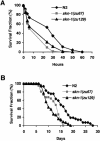
Figures
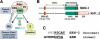

References
-
- Blackwell T.K., Bowerman, B., Priess, J., and Weintraub, H. 1994. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science 266: 621-628. - PubMed
-
- Bluher M., Kahn, B.B., and Kahn, C.R. 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299: 572-574. - PubMed
-
- Bowerman B., Eaton, B.A., and Priess, J.R. 1992. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68: 1061-1075. - PubMed
-
- Bowerman B., Draper, B.W., Mello, C., and Priess, J. 1993. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74: 443-452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
