Single-molecule transition-state analysis of RNA folding
- PMID: 12869691
- PMCID: PMC170913
- DOI: 10.1073/pnas.1133280100
Single-molecule transition-state analysis of RNA folding
Abstract
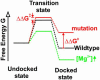
How RNA molecules fold into functional structures is a problem of great significance given the expanding list of essential cellular RNA enzymes and the increasing number of applications of RNA in biotechnology and medicine. A critical step toward solving the RNA folding problem is the characterization of the associated transition states. This is a challenging task in part because the rugged energy landscape of RNA often leads to the coexistence of multiple distinct structural transitions. Here, we exploit single-molecule fluorescence spectroscopy to follow in real time the equilibrium transitions between conformational states of a model RNA enzyme, the hairpin ribozyme. We clearly distinguish structural transitions between effectively noninterchanging sets of unfolded and folded states and characterize key factors defining the transition state of an elementary folding reaction where the hairpin ribozyme's two helical domains dock to make several tertiary contacts. Our single-molecule experiments in conjunction with site-specific mutations and metal ion titrations show that the two RNA domains are in a contact or close-to-contact configuration in the transition state even though the native tertiary contacts are at most partially formed. Such a compact transition state without well formed tertiary contacts may be a general property of elementary RNA folding reactions.
Figures
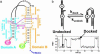
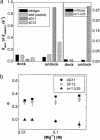
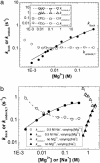

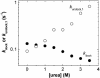
References
-
- Doudna, J. A. & Cech, T. R. (2002) Nature 418, 222–228. - PubMed
-
- Moore, P. B. & Steitz, T. A. (2002) Nature 418, 229–235. - PubMed
-
- Collins, C. A. & Guthrie, C. (2000) Nat. Struct. Biol. 7, 850–854. - PubMed
-
- Valadkhan, S. & Manley, J. L. (2002) Nat. Struct. Biol. 9, 498–499. - PubMed
-
- Famulok, M. & Verma, S. (2002) Trends Biotechnol. 20, 462–466. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

