Prediction of clinical drug efficacy by classification of drug-induced genomic expression profiles in vitro
- PMID: 12869696
- PMCID: PMC170965
- DOI: 10.1073/pnas.1632587100
Prediction of clinical drug efficacy by classification of drug-induced genomic expression profiles in vitro
Abstract
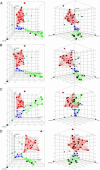
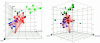
Assays of drug action typically evaluate biochemical activity. However, accurately matching therapeutic efficacy with biochemical activity is a challenge. High-content cellular assays seek to bridge this gap by capturing broad information about the cellular physiology of drug action. Here, we present a method of predicting the general therapeutic classes into which various psychoactive drugs fall, based on high-content statistical categorization of gene expression profiles induced by these drugs. When we used the classification tree and random forest supervised classification algorithms to analyze microarray data, we derived general "efficacy profiles" of biomarker gene expression that correlate with anti-depressant, antipsychotic and opioid drug action on primary human neurons in vitro. These profiles were used as predictive models to classify naïve in vitro drug treatments with 83.3% (random forest) and 88.9% (classification tree) accuracy. Thus, the detailed information contained in genomic expression data is sufficient to match the physiological effect of a novel drug at the cellular level with its clinical relevance. This capacity to identify therapeutic efficacy on the basis of gene expression signatures in vitro has potential utility in drug discovery and drug target validation.
Figures
References
-
- Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109-126. - PubMed
-
- Pomeroy, S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M., Angelo, M., McLaughlin, M. E., Kim, J. Y., Goumnerova, L. C., Black, P. M., Lau, C., et al. (2002) Nature 415, 436-442. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

