Ribozyme speed limits
- PMID: 12869701
- PMCID: PMC1370456
- DOI: 10.1261/rna.5680603
Ribozyme speed limits
Abstract
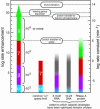
The speed at which RNA molecules decompose is a critical determinant of many biological processes, including those directly involved in the storage and expression of genetic information. One mechanism for RNA cleavage involves internal phosphoester transfer, wherein the 2'-oxygen atom carries out an SN2-like nucleophilic attack on the adjacent phosphorus center (transesterification). In this article, we discuss fundamental principles of RNA transesterification and define a conceptual framework that can be used to assess the catalytic power of enzymes that cleave RNA. We deduce that certain ribozymes and deoxyribozymes, like their protein enzyme counterparts, can bring about enormous rate enhancements.
Figures
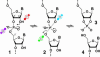


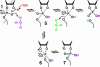

References
-
- Adams, R.L.P., Knowler, J.T., and Leader, D.P. 1992. Degradation and modification of nucleic acids. In The biochemistry of the nucleic acids, 11th ed., pp. 97–133. Chapman & Hall, New York.
-
- Admiraal, S.J. and Herschlag, D. 1995. Mapping the transition state for ATP hydrolysis: Implications for enzymatic catalysis. Chem. Biol. 2: 729–739. - PubMed
-
- Almer, H. and Strömberg, R. 1996. Base catalysis and leaving group dependence in intramolecular alcoholysis of uridine 3′-(aryl phosphorothioate)s. J. Am. Chem. Soc. 118: 7921–7928.
-
- Bacher, J.E. and Kauzmann, W. 1952. The kinetics of hydrolysis of ribonucleic acid. J. Am. Chem. Soc. 74: 3779–3786.
-
- Benkovic, S.J. and Schray, K.J. 1970. Chemical basis of biological phosphoryl transfer. In The enzymes, 3rd ed., Vol. VIII (ed. P.D. Boyer), pp. 201–238, Academic Press, New York.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
