A common speed limit for RNA-cleaving ribozymes and deoxyribozymes
- PMID: 12869706
- PMCID: PMC1370461
- DOI: 10.1261/rna.5670703
A common speed limit for RNA-cleaving ribozymes and deoxyribozymes
Abstract
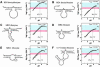
It is widely believed that the reason proteins dominate biological catalysis is because polypeptides have greater chemical complexity compared with nucleic acids, and thus should have greater enzymatic power. Consistent with this hypothesis is the fact that protein enzymes typically exhibit chemical rate enhancements that are far more substantial than those achieved by natural and engineered ribozymes. To investigate the true catalytic power of nucleic acids, we determined the kinetic characteristics of 14 classes of engineered ribozymes and deoxyribozymes that accelerate RNA cleavage by internal phosphoester transfer. Half approach a maximum rate constant of approximately 1 min(-1), whereas ribonuclease A catalyzes the same reaction approximately 80,000-fold faster. Additional biochemical analyses indicate that this commonly encountered ribozyme "speed limit" coincides with the theoretical maximum rate enhancement for an enzyme that uses only two specific catalytic strategies. These results indicate that ribozymes using additional catalytic strategies could be made that promote RNA cleavage with rate enhancements that equal those of proteins.
Figures
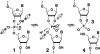



References
-
- Breaker, R.R. 1997. In vitro selection of catalytic polynucleotides. Chem. Rev. 97: 371–390. - PubMed
-
- Burgers, P.M.J. and Eckstein, F. 1979. Diastereomers of 5′-O-adenosyl 3′-O-uridyl phosphorothioate: Chemical synthesis and enzymatic properties. Biochemistry 18: 592–596. - PubMed
-
- Butcher, S.E. 2001. Structure and function of the small ribozymes. Curr. Opin. Struct. Biol. 11: 315–320. - PubMed
-
- Carola, C. and Eckstein, F. 1999. Nucleic acid enzymes. Curr. Opin. Chem. Biol. 3: 274–283. - PubMed
-
- Clouet-d’Orval, B. and Uhlenbeck, O.C. 1997. Hammerhead ribozymes with a faster cleavage rate. Biochemistry 36: 9087–9092. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
