Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae
- PMID: 12869710
- PMCID: PMC1370465
- DOI: 10.1261/rna.5390803
Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae
Abstract
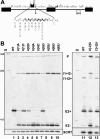
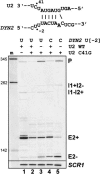
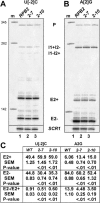
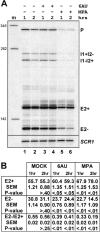
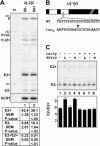
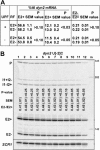
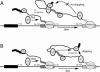
Unknown mechanisms exist to ensure that exons are not skipped during biogenesis of mRNA. Studies have connected transcription elongation with regulated alternative exon inclusion. To determine whether the relative rates of transcription elongation and spliceosome assembly might play a general role in enforcing constitutive exon inclusion, we measured exon skipping for a natural two-intron gene in which the internal exon is constitutively included in the mRNA. Mutations in this gene that subtly reduce recognition of the intron 1 branchpoint cause exon skipping, indicating that rapid recognition of the first intron is important for enforcing exon inclusion. To test the role of transcription elongation, we treated cells to increase or decrease the rate of transcription elongation. Consistent with the "first come, first served" model, we found that exon skipping in vivo is inhibited when transcription is slowed by RNAP II mutants or when cells are treated with inhibitors of elongation. Expression of the elongation factor TFIIS stimulates exon skipping, and this effect is eliminated when lac repressor is targeted to DNA encoding the second intron. A mutation in U2 snRNA promotes exon skipping, presumably because a delay in recognition of the first intron allows elongating RNA polymerase to transcribe the downstream intron. This indicates that the relative rates of elongation and splicing are tuned so that the fidelity of exon inclusion is enhanced. These findings support a general role for kinetic coordination of transcription elongation and splicing during the transcription-dependent control of splicing.
Figures

References
-
- Abovich, N. and Rosbash, M. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412. - PubMed
-
- Abovich, N., Liao, X.C., and Rosbash, M. 1994. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes & Dev. 8: 843–854. - PubMed
-
- Aebi, M. and Weissman, S.M. 1987. Precision and orderliness in splicing. Trends Genet. 3: 102–107.
-
- Ares Jr., M. and Igel, A.H. 1990. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes & Dev. 4: 2132–2145. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
