A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA
- PMID: 12869711
- PMCID: PMC1370466
- DOI: 10.1261/rna.5590603
A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA
Abstract
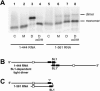
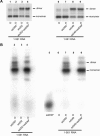
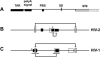
The 5' untranslated leader region of retroviral RNAs contains noncoding information that is essential for viral replication, including signals for transcriptional transactivation, splicing, primer binding for reverse transcription, dimerization of the genomic RNA, and encapsidation of the viral RNA into virions. These RNA motifs have considerable structural and functional overlap. In this study, we investigate the conformational dynamics associated with the use and silencing of a sequence in HIV-2 RNA that is involved in genomic RNA dimerization called stem-loop 1 (SL1) and its relationship with a flanking sequence that is known to be important for encapsidation of viral RNAs. We demonstrate that a long-distance intramolecular interaction between nucleotides located upstream of the primer-binding site domain and nucleotides encompassing the Gag translation start codon functionally silences SL1 as a dimerization element. This silencing can be relieved by mutation or by hybridization of an oligonucleotide that disrupts the long-distance interaction. Furthermore, we identify a palindrome within the packaging/encapsidation signal Psi (just 5' of SL1) that can either serve as an efficient dimerization signal itself, or can mediate SL1 silencing through base pairing with SL1. These results provide a tangible link between the functions of genomic RNA dimerization and encapsidation, which are known to be related, but whose physical relationship has been unclear. A model is proposed that accounts for observations of dimerization, packaging, and translation of viral RNAs during different phases of the viral replication cycle.
Figures



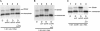


Similar articles
-
Regulation of primate lentiviral RNA dimerization by structural entrapment.Retrovirology. 2008 Jul 17;5:65. doi: 10.1186/1742-4690-5-65. Retrovirology. 2008. PMID: 18637186 Free PMC article.
-
Elements located upstream and downstream of the major splice donor site influence the ability of HIV-2 leader RNA to dimerize in vitro.Biochemistry. 2003 Mar 11;42(9):2634-42. doi: 10.1021/bi0271190. Biochemistry. 2003. PMID: 12614158 Free PMC article.
-
HIV-2 RNA dimerization is regulated by intramolecular interactions in vitro.RNA. 2007 Aug;13(8):1341-54. doi: 10.1261/rna.483807. Epub 2007 Jun 25. RNA. 2007. PMID: 17592043 Free PMC article.
-
Dimerization of retroviral genomic RNAs: structural and functional implications.Biochimie. 1996;78(7):639-53. doi: 10.1016/s0300-9084(96)80010-1. Biochimie. 1996. PMID: 8955907 Review.
-
Retroviral RNA packaging: a review.Arch Virol Suppl. 1994;9:513-22. doi: 10.1007/978-3-7091-9326-6_49. Arch Virol Suppl. 1994. PMID: 8032280 Review.
Cited by
-
The matrix domain contributes to the nucleic acid chaperone activity of HIV-2 Gag.Retrovirology. 2016 Mar 17;13:18. doi: 10.1186/s12977-016-0245-1. Retrovirology. 2016. PMID: 26987314 Free PMC article.
-
Splicing affects presentation of RNA dimerization signals in HIV-2 in vitro.Nucleic Acids Res. 2004 Aug 27;32(15):4585-95. doi: 10.1093/nar/gkh800. Print 2004. Nucleic Acids Res. 2004. PMID: 15333691 Free PMC article.
-
SHAPE analysis of the 5' end of the Mason-Pfizer monkey virus (MPMV) genomic RNA reveals structural elements required for genome dimerization.RNA. 2013 Dec;19(12):1648-58. doi: 10.1261/rna.040931.113. Epub 2013 Oct 23. RNA. 2013. PMID: 24152551 Free PMC article.
-
Retroviral RNA Dimerization: From Structure to Functions.Front Microbiol. 2018 Mar 22;9:527. doi: 10.3389/fmicb.2018.00527. eCollection 2018. Front Microbiol. 2018. PMID: 29623074 Free PMC article. Review.
-
Structural basis of genomic RNA (gRNA) dimerization and packaging determinants of mouse mammary tumor virus (MMTV).Retrovirology. 2014 Nov 14;11:96. doi: 10.1186/s12977-014-0096-6. Retrovirology. 2014. PMID: 25394412 Free PMC article.
References
-
- Abbink, T.E. and Berkhout, B. 2002. A novel long distance base pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 278: 11601–11611. - PubMed
-
- Bender, W. and Davidson, N. 1976. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell 7: 595–607. - PubMed
-
- Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54: 1–34. - PubMed
-
- Berkhout, B., Ooms, M., Beerens, N., Huthoff, H., Southern, E., and Verhoef, K. 2002. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 277: 19967–19975. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
