When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism
- PMID: 12874385
- PMCID: PMC187858
- DOI: 10.1073/pnas.1632156100
When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism
Abstract
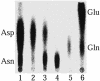
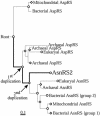
Faithful protein synthesis relies on a family of essential enzymes called aminoacyl-tRNA synthetases, assembled in a piecewise fashion. Analysis of the completed archaeal genomes reveals that all archaea that possess asparaginyl-tRNA synthetase (AsnRS) also display a second ORF encoding an AsnRS truncated from its anticodon binding-domain (AsnRS2). We show herein that Pyrococcus abyssi AsnRS2, in contrast to AsnRS, does not sustain asparaginyl-tRNAAsn synthesis but is instead capable of converting aspartic acid into asparagine. Functional analysis and complementation of an Escherichia coli asparagine auxotrophic strain show that AsnRS2 constitutes the archaeal homologue of the bacterial ammonia-dependent asparagine synthetase A (AS-A), therefore named archaeal asparagine synthetase A (AS-AR). Primary sequence- and 3D-based phylogeny shows that an archaeal AspRS ancestor originated AS-AR, which was subsequently transferred into bacteria by lateral gene transfer in which it underwent structural changes producing AS-A. This study provides evidence that a contemporary aminoacyl-tRNA synthetase can be recruited to sustain amino acid metabolism.
Figures
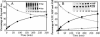

Comment in
-
tRNA synthetase paralogs: evolutionary links in the transition from tRNA-dependent amino acid biosynthesis to de novo biosynthesis.Proc Natl Acad Sci U S A. 2003 Aug 19;100(17):9650-2. doi: 10.1073/pnas.1934245100. Epub 2003 Aug 11. Proc Natl Acad Sci U S A. 2003. PMID: 12913115 Free PMC article. No abstract available.
Similar articles
-
Crystal structure of the archaeal asparagine synthetase: interrelation with aspartyl-tRNA and asparaginyl-tRNA synthetases.J Mol Biol. 2011 Sep 23;412(3):437-52. doi: 10.1016/j.jmb.2011.07.050. Epub 2011 Jul 28. J Mol Biol. 2011. PMID: 21820443
-
Structural basis of the water-assisted asparagine recognition by asparaginyl-tRNA synthetase.J Mol Biol. 2006 Jul 7;360(2):329-42. doi: 10.1016/j.jmb.2006.04.068. Epub 2006 May 15. J Mol Biol. 2006. PMID: 16753178
-
Glu-Q-tRNA(Asp) synthetase coded by the yadB gene, a new paralog of aminoacyl-tRNA synthetase that glutamylates tRNA(Asp) anticodon.Biochimie. 2005 Sep-Oct;87(9-10):847-61. doi: 10.1016/j.biochi.2005.03.007. Epub 2005 Apr 8. Biochimie. 2005. PMID: 16164993
-
Substrate selection by aminoacyl-tRNA synthetases.Nucleic Acids Symp Ser. 1995;(33):40-2. Nucleic Acids Symp Ser. 1995. PMID: 8643392 Review.
-
Aminoacyl-tRNA synthesis by pre-translational amino acid modification.RNA Biol. 2004 May;1(1):16-20. Epub 2004 May 28. RNA Biol. 2004. PMID: 17194933 Review.
Cited by
-
Turning tRNA upside down: When aminoacylation is not a prerequisite to protein synthesis.Proc Natl Acad Sci U S A. 2004 May 18;101(20):7493-4. doi: 10.1073/pnas.0402276101. Epub 2004 May 11. Proc Natl Acad Sci U S A. 2004. PMID: 15138304 Free PMC article. No abstract available.
-
PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica.Mol Cell. 2010 Jul 30;39(2):209-21. doi: 10.1016/j.molcel.2010.06.021. Mol Cell. 2010. PMID: 20670890 Free PMC article.
-
Plasmodium Apicoplast Gln-tRNAGln Biosynthesis Utilizes a Unique GatAB Amidotransferase Essential for Erythrocytic Stage Parasites.J Biol Chem. 2015 Dec 4;290(49):29629-41. doi: 10.1074/jbc.M115.655100. Epub 2015 Aug 28. J Biol Chem. 2015. PMID: 26318454 Free PMC article.
-
Coevolution Theory of the Genetic Code at Age Forty: Pathway to Translation and Synthetic Life.Life (Basel). 2016 Mar 16;6(1):12. doi: 10.3390/life6010012. Life (Basel). 2016. PMID: 26999216 Free PMC article.
-
An unusual tryptophanyl tRNA synthetase interacts with nitric oxide synthase in Deinococcus radiodurans.Proc Natl Acad Sci U S A. 2004 Nov 9;101(45):15881-6. doi: 10.1073/pnas.0405483101. Epub 2004 Nov 1. Proc Natl Acad Sci U S A. 2004. PMID: 15520379 Free PMC article.
References
-
- Ibba, M., Francklyn, C. & Cusack, S., eds. (2003) The Aminoacyl-tRNA Synthetases (Landes Bioscience, Georgetown, TX), in press.
-
- Ibba, M., Becker, H. D., Stathopoulos, C., Tumbula, D. L. & Söll, D. (2000) Trends Biochem. Sci. 25, 311–316. - PubMed
-
- Curnow, A. W., Ibba, M. & Söll, D. (1996) Nature 382, 589–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

