A chemical approach for identifying O-GlcNAc-modified proteins in cells
- PMID: 12874386
- PMCID: PMC171382
- DOI: 10.1073/pnas.1632821100
A chemical approach for identifying O-GlcNAc-modified proteins in cells
Abstract
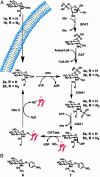
The glycosylation of serine and threonine residues with a single GlcNAc moiety is a dynamic posttranslational modification of many nuclear and cytoplasmic proteins. We describe a chemical strategy directed toward identifying O-GlcNAc-modified proteins from living cells or proteins modified in vitro. We demonstrate, in vitro, that each enzyme in the hexosamine salvage pathway, and the enzymes that affect this dynamic modification (UDP-GlcNAc:polypeptidtyltransferase and O-GlcNAcase), tolerate analogues of their natural substrates in which the N-acyl side chain has been modified to bear a bio-orthogonal azide moiety. Accordingly, treatment of cells with N-azidoacetylglucosamine results in the metabolic incorporation of the azido sugar into nuclear and cytoplasmic proteins. These O-azidoacetylglucosamine-modified proteins can be covalently derivatized with various biochemical probes at the site of protein glycosylation by using the Staudinger ligation. The approach was validated by metabolic labeling of nuclear pore protein p62, which is known to be posttranslationally modified with O-GlcNAc. This strategy will prove useful for both the identification of O-GlcNAc-modified proteins and the elucidation of the specific residues that bear this saccharide.
Figures
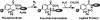


References
-
- Torres, C. R. & Hart, G. W. (1984) J. Biol. Chem. 259, 3308–3317. - PubMed
-
- Wells, L., Vosseller, K. & Hart, G. W. (2001) Science 291, 2376–2378. - PubMed
-
- Hanover, J. A. (2001) FASEB J. 15, 1865–1876. - PubMed
-
- Zachara, N. E. & Hart, G. W. (2002) Chem. Rev. 102, 431–438. - PubMed
-
- Jackson, S. P. & Tjian, R. (1988) Cell 55, 125–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

