Memory in retroviral quasispecies: experimental evidence and theoretical model for human immunodeficiency virus
- PMID: 12875847
- PMCID: PMC7173031
- DOI: 10.1016/s0022-2836(03)00661-2
Memory in retroviral quasispecies: experimental evidence and theoretical model for human immunodeficiency virus
Abstract
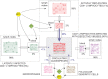
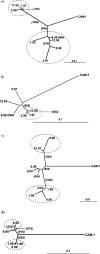
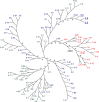
Viral quasispecies may possess a molecular memory of their past evolutionary history, imprinted on minority components of the mutant spectrum. Here we report experimental evidence and a theoretical model for memory in retroviral quasispecies in vivo. Apart from replicative memory associated with quasispecies dynamics, retroviruses may harbour a "cellular" or "anatomical" memory derived from their integrative cycle and the presence of viral reservoirs in body compartments. Three independent sets of data exemplify the two kinds of memory in human immunodeficiency virus type 1 (HIV-1). The data provide evidence of re-emergence of sequences that were hidden in cellular or anatomical compartments for extended periods of infection, and recovery of a quasispecies from pre-existing genomes. We develop a three-component model that incorporates the essential features of the quasispecies dynamics of retroviruses exposed to selective pressures. Significantly, a numerical study based on this model is in agreement with the experimental data, further supporting the existence of both replicative and reservoir memory in retroviral quasispecies.
Figures

References
-
- Eigen M., Biebricher C.K. Sequence space and quasispecies distribution. In: Domingo E., Ahlquist P., Holland J.J., editors. vol. 3. CRC Press; Boca Raton, FL: 1988. (RNA Genetics).
-
- Eigen M. On the nature of virus quasispecies. Trends Microbiol. 1996;4:216–218. - PubMed
-
- Domingo E., Biebricher C., Holland J.J., Eigen M. Quasispecies and RNA Virus Evolution: Principles and Consequences. Landes; Austin, TX: 2001.
-
- Eigen M. Self-organization of matter and evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

